BioBlend API
The Galaxy API enables developers to access Galaxy functionalities using high-level scripts. BioBlend is a Python overlay implemented makes it easy for bioinformaticians to automate end-to-end large data analysis, from scratch, in a way that is highly accessible to collaborators, by allowing them to both provide the required infrastructure and automate complex analyses over large datasets within the familiar Galaxy environment.
Links
- Source code for bioblend.galaxy
- API documentation
- On github
- Virtualenv
- Jupyter Notebook
- BioBlend Tutorial
Galaxy API
The web interface is appropriate for things like exploratory analysis, visualization, construction of workflows, and rerunning workflows on new datasets. It is thus an excellent tool for designing analyses, recording provenance and facilitating collaboration between bioinformaticians and biologists. But the web interface is less suitable for things like:
- Connecting a Galaxy instance directly to your sequencer and running workflows whenever data is ready
- Running a workflow against multiple datasets (which can be done with the web interface, but is tedious)
- When the analysis involves complex control, such as looping and branching.
The Galaxy API addresses these and other situations by exposing Galaxy internals through an additional interface, known as an Applications Programming Interface, or API.
Enabling API
To use the API, you must first generate an API Key for the account you want to access Galaxy from. You can do so in the UI under user preferences (while logged in).
Programming Language Bindings
Various language specific libraries for interfacing with the Galaxy API have been developed by the Galaxy community:
- The Galaxy code itself contains JavaScript bindings.
- BioBlend contains Python bindings:
- blend4j contains Java bindings largely modeled after BioBlend.
- blend4php contains PHP bindings.
BioBlend Overview
BioBlend is a Python library implemented to interact with Galaxy via a straightforward API.
BioBlend enable developers to manipulate genomic analyses within Galaxy, including data management and workflow execution.
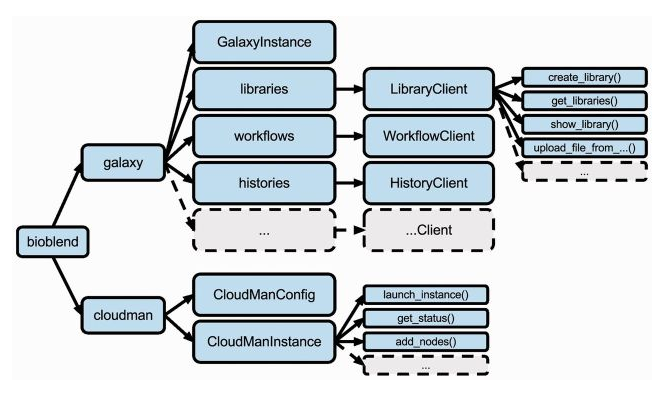 An alternative object-oriented API is also available, but it’s still incomplete, providing access to a more restricted set of Galaxy modules with respect to the standard one.
An alternative object-oriented API is also available, but it’s still incomplete, providing access to a more restricted set of Galaxy modules with respect to the standard one.
Installation
Stable releases of BioBlend are best installed via pip or easy_install.
pip install bioblend
Alternatively, you may install the most current source code from Git repository.
# Clone the repository to a local directory
$ git clone https://github.com/galaxyproject/bioblend.git
# Install the library
$ cd bioblend
$ python setup.py install
After installing the library, you will be able to simply import it into your Python environment with:
import bioblend
GalaxyInstance
A base representation of an instance of Galaxy, identified by a URL and a user’s API key.
from bioblend import galaxy
gi = galaxy.GalaxyInstance(url='http://127.0.0.1:8000', key='your_api_key')
After you have created an GalaxyInstance object, access various modules via the class fields : config, libraries, histories, workflows, datasets, and users
config = gi.config.get_config()
users = gi.users.get_users()
histories = gi.histories.get_histories()
Config
Contains possible interaction dealing with Galaxy configuration. (View source)
class bioblend.galaxy.config.ConfigClient(galaxy_instance)
get_config()
Get a list of attributes about the Galaxy instance. More attributes will be present if the user is an admin.
config = gi.config.get_config()
Return type: list
Returns: A list of attributes.
{u'allow_library_path_paste': False,
u'allow_user_creation': True,
u'allow_user_dataset_purge': True,
u'allow_user_deletion': False,
u'enable_unique_workflow_defaults': False,
u'ftp_upload_dir': u'/SOMEWHERE/galaxy/ftp_dir',
u'ftp_upload_site': u'galaxy.com',
u'library_import_dir': u'None',
u'logo_url': None,
u'support_url': u'https://galaxyproject.org/support',
u'terms_url': None,
u'user_library_import_dir': None,
u'wiki_url': u'https://galaxyproject.org/'}
get_version()
Get the current version of the Galaxy instance.
version = gi.config.get_version()
Return type: dict
Returns: Version of the Galaxy instance
{u'extra': {}, u'version_major': u'17.05'}
Libraries
Contains possible interactions with the Galaxy Data Libraries (View source)
class bioblend.galaxy.libraries.LibraryClient(galaxy_instance)
get_libraries()
Get all the libraries or filter for specific one(s) via the provided name or ID.
libraries = gi.libraries.get_libraries(library_id=None, name=None, deleted=False)
Provide only one argument: name or library_id, but not both.
If deleted is set to True, return libraries that have been deleted.
Return type: List
Returns: list of dicts each containing basic information about a library
[{u'can_user_add': False,
u'can_user_manage': False,
u'can_user_modify': False,
u'create_time': u'2016-12-08T10:55:24.905561',
u'create_time_pretty': u'6 months ago',
u'deleted': False,
u'description': u'',
u'id': u'105bb2d5978eda3f',
u'model_class': u'Library',
u'name': u'first private library',
u'public': True,
u'root_folder_id': u'Fb2598cee3751a874',
u'synopsis': u''},
{u'can_user_add': False,
u'can_user_manage': False,
u'can_user_modify': False,
u'create_time': u'2009-07-30T15:11:06.232324',
u'create_time_pretty': u'7 years ago',
u'deleted': True,
u'description': u'',
u'id': u'2498224736779bdb',
u'model_class': u'Library',
u'name': u'Nate Test',
u'public': True,
u'root_folder_id': u'F652f5dd81838e917',
u'synopsis': None}]
show_library()
Get information about a library, by library_id
library = gi.libraries.show_library(library_id, contents=False)
contents=True if want to get contents of the library (rather than just the library details)
Return type: Dict
Returns: Dictionary containg details of the given library
# id contents=False
{u'can_user_add': False,
u'can_user_manage': False,
u'can_user_modify': False,
u'create_time': u'2016-12-08T10:55:24.905561',
u'create_time_pretty': u'6 months ago',
u'deleted': False,
u'description': u'',
u'id': u'105bb2d5978eda3f',
u'model_class': u'Library',
u'name': u'first private library',
u'public': True,
u'root_folder_id': u'Fb2598cee3751a874',
u'synopsis': u''}
# if contents=True
[{u'id': u'Fb2598cee3751a874',
u'name': u'/',
u'type': u'folder',
u'url': u'/api/libraries/105bb2d5978eda3f/contents/Fb2598cee3751a874'}]
create_library()
Create a data library with the properties defined in the arguments.
l = gi.libraries.create_library(name, description=None, synopsis=None)
Return type: Dict
Returns: Details of the created library.
{u'can_user_add': False,
u'can_user_manage': False,
u'can_user_modify': False,
u'create_time': u'2017-06-20T10:40:07.203839',
u'create_time_pretty': u'',
u'deleted': False,
u'description': u'Library from bioblend',
u'id': u'af2dc2a1ab633018',
u'model_class': u'Library',
u'name': u'Dummy Library',
u'public': True,
u'root_folder_id': u'F6e353dd7bf64a026',
u'synopsis': u''}
delete_library()
Delete a data library
l = gi.libraries.delete_library(library_id)
get_folders()
Get all the folders or filter specific one(s) via the provided name or folder_id in data library with id library_id.
folders = gi.libraries.get_folders(library_id, folder_id=None, name=None)
Provide only one argument: name or folder_id, but not both.
For name specify the full path of the folder starting from the library’s root folder, e.g. /subfolder/subsubfolder.
Return type: List
Returns: list of dicts each containing basic information about a folder
[{u'id': u'Fb2598cee3751a874',
u'name': u'/',
u'type': u'folder',
u'url': u'/api/libraries/105bb2d5978eda3f/contents/Fb2598cee3751a874'}]
show_folder()
Get details about a given folder.
folder = gi.libraries.show_folder(library_id, folder_id)
The required folder_id can be obtained from the folder’s library content details.
create_folder()
Create a folder in a library
folder = gi.libraries.create_folder(library_id, folder_name, description=None, base_folder_id=None)
base_folder_id is the id of the folder where to create the new folder. If not provided, the root folder will be used
Return type: Dict
Returns: Dictionary each containing basic information about created folder
[{u'id': u'F2f38cd1ede810ca1',
u'name': u'Dummy Folder',
u'url': u'/api/libraries/af2dc2a1ab633018/contents/F2f38cd1ede810ca1'}]
Datasets in Libraries
copy_from_dataset()
Copy a Galaxy dataset into a library.
gi.libraries.copy_from_dataset(library_id, dataset_id, folder_id=None, message='')
forder_id:the id of the folder where to place the uploaded files. If not provided, the root folder will be used.message: message note for copying action
upload_file_contents()
Upload pasted_content to a data library as a new file.
gi.libraries.upload_file_contents(library_id, pasted_content, folder_id=None, file_type='auto', dbkey='?')
pasted_content(str) – Content to upload into the libraryfolder_id(str) – id of the folder where to place the uploaded file. If not provided, the root folder will be usedfile_type(str) – Galaxy file format namedbkey(str) – Dbkey
upload_file_from_local_path()
Read local file contents from file_local_path and upload data to a given library.
gi.libraries.upload_file_from_local_path(library_id, file_local_path, folder_id=None, file_type='auto', dbkey='?')
file_local_path(str) – path of local file to uploadfolder_id(str) – id of the folder where to place the uploaded file. If not provided, the root folder will be usedfile_type(str) – Galaxy file format namedbkey(str) – Dbkey
upload_file_from_server()
Upload all files in the specified subdirectory of the Galaxy library import directory to a given library.
gi.libraries.upload_file_from_server(library_id, server_dir, folder_id=None, file_type='auto', dbkey='?', link_data_only=None, roles='')
For this method to work, the Galaxy instance must have the library_import_dir option configured in the config/galaxy.ini configuration file.
server_dir(str) – relative path of the subdirectory of library_import_dir to upload. All and only the files (i.e. no subdirectories) contained in the specified directory will be uploadedfolder_id(str) – id of the folder where to place the uploaded files. If not provided, the root folder will be usedfile_type(str) – Galaxy file format namedbkey(str) – Dbkeylink_data_only(str) – either ‘copy_files’ (default) or ‘link_to_files’. Setting to ‘link_to_files’ symlinks instead of copying the filesroles(str) – ???
upload_file_from_url()
Upload a file to a given library from a URL.
gi.libraries.upload_file_from_url(library_id, file_url, folder_id=None, file_type='auto', dbkey='?')
file_url(str) – URL of the file to uploadfolder_id(str) – id of the folder where to place the uploaded files. If not provided, the root folder will be usedfile_type(str) – Galaxy file format namedbkey(str) – Dbkey
upload_from_galaxy_filesystem()
Upload a set of files already present on the filesystem of the Galaxy server to a given library.
gi.libraries.upload_from_galaxy_filesystem(library_id, filesystem_paths, folder_id=None, file_type='auto', dbkey='?', link_data_only=None, roles='')
For this method to work, the Galaxy instance must have the allow_library_path_paste option set to True in the config/galaxy.ini configuration file.
filesystem_paths(str) – file paths on the Galaxy server to upload to the library, one file per linefolder_id(str) – id of the folder where to place the uploaded files. If not provided, the root folder will be usedfile_type(str) – Galaxy file format namedbkey(str) – Dbkeylink_data_only(str) – either ‘copy_files’ (default) or ‘link_to_files’. Setting to ‘link_to_files’ symlinks instead of copying the filesroles(str) – ???
delete_library_dataset()
Delete a library dataset in a data library (Mark it as deleted)
d = gi.libraries.delete_library_dataset(library_id, dataset_id, purged=False)
if purge=True, also purge (permanently delete) the dataset.
Return type: dict
Returns: A dictionary containing the dataset id and whether the dataset has been deleted.
{u'deleted': True,
u'id': u'60e680a037f41974'}
Examples
Histories
Contains possible interactions with the Galaxy Histories (View source)
class bioblend.galaxy.histories.HistoryClient(galaxy_instance)
get_histories()
Get all histories or filter the specific one(s) via the provided name or history_id.
histories = gi.histories.get_histories(history_id=None, name=None, deleted=False)
Provide only one argument, name or history_id, but not both.
If deleted is set to True, return histories that have been deleted.
Return type: list
Returns: A list of history element dicts. If more than one history matches the givenname, return the list of all the histories with the given name
[{u'annotation': None,
u'deleted': False,
u'id': u'7f929922b23b572f',
u'model_class': u'History',
u'name': u'Dummy history',
u'published': False,
u'purged': False,
u'tags': [],
u'url': u'/api/histories/7f929922b23b572f'}]
show_history()
Get details of a given history. By default, just get the history meta information.
h = gi.histories.show_history(history_id, contents=False, deleted=None, visible=None, details=None, types=None)
When contents=True, show_histor() returns the complete list of datasets in the given history:
deleted: includes deleted datasets in history dataset listvisible: includes only visible datasets in history dataset listdetails: includes dataset details. Set to ‘all’ for the most informationReturn type: dict
Returns: details of the given history
{u'annotation': None,
u'contents_url': u'/api/histories/7f929922b23b572f/contents',
u'create_time': u'2017-01-10T13:28:03.575271',
u'deleted': False,
u'empty': False,
u'genome_build': u'?',
u'id': u'7f929922b23b572f',
u'importable': False,
u'model_class': u'History',
u'name': u'Dummy history',
u'published': False,
u'purged': False,
u'size': 408,
u'slug': None,
u'state': u'queued',
u'state_details': {u'discarded': 0,
u'empty': 0,
u'error': 4,
u'failed_metadata': 0,
u'new': 6,
u'ok': 5,
u'paused': 3,
u'queued': 0,
u'running': 0,
u'setting_metadata': 0,
u'upload': 0},
u'state_ids': {u'discarded': [],
u'empty': [],
u'error': [u'bbd44e69cb8906b55d65844668967e13',
u'bbd44e69cb8906b54c7a94ab2e8141db',
u'bbd44e69cb8906b5cc2a51c1c19368b5',
u'bbd44e69cb8906b54ce551cbcb48857f'],
u'failed_metadata': [],
u'new': [u'bbd44e69cb8906b5c903f9b2ad625329',
u'bbd44e69cb8906b5400483ab6b6db0bf',
u'bbd44e69cb8906b5ed51476b38eb4011',
u'bbd44e69cb8906b50ab8299b1973e8aa',
u'bbd44e69cb8906b5c4d0a26e6a56f9bf',
u'bbd44e69cb8906b58de2f1230120bae4'],
u'ok': [u'bbd44e69cb8906b5ce7fe31bd9b0016a',
u'bbd44e69cb8906b5a90837cc6d7119fb',
u'bbd44e69cb8906b5da3212bbb1d5b2ad',
u'bbd44e69cb8906b5c6d64c61a117ad23',
u'bbd44e69cb8906b5a1e757c207380ab5',
u'bbd44e69cb8906b5f12e91ed91c973e8',
u'bbd44e69cb8906b5792974169f04a06f',
u'bbd44e69cb8906b5b11e71f4d1889ae2'],
u'paused': [u'bbd44e69cb8906b5e4c8e2714eb61373',
u'bbd44e69cb8906b5933289da7d7c112b',
u'bbd44e69cb8906b580708c09b4073ac7'],
u'queued': [],
u'running': [],
u'setting_metadata': [],
u'upload': []},
u'tags': [],
u'update_time': u'2017-01-13T14:30:36.179669',
u'url': u'/api/histories/7f929922b23b572f',
u'user_id': u'bd0631c5dfef81b1',
u'username_and_slug': None}
create_history()
Create a new history, optionally setting the name.
h = gi.histories.create_history(name=None)
Return type: dict
Returns: Dictionary containing information about newly created history
{u'annotation': None,
u'contents_url': u'/api/histories/a956ab1d3d21799b/contents',
u'create_time': u'2017-06-19T16:45:36.035290',
u'deleted': False,
u'empty': True,
u'genome_build': None,
u'id': u'a956ab1d3d21799b',
u'importable': False,
u'model_class': u'History',
u'name': u'Unnamed history',
u'published': False,
u'purged': False,
u'size': 0,
u'slug': None,
u'state': u'new',
u'state_details': {u'discarded': 0,
u'empty': 0,
u'error': 0,
u'failed_metadata': 0,
u'new': 0,
u'ok': 0,
u'paused': 0,
u'queued': 0,
u'running': 0,
u'setting_metadata': 0,
u'upload': 0},
u'state_ids': {u'discarded': [],
u'empty': [],
u'error': [],
u'failed_metadata': [],
u'new': [],
u'ok': [],
u'paused': [],
u'queued': [],
u'running': [],
u'setting_metadata': [],
u'upload': []},
u'tags': [],
u'update_time': u'2017-06-19T16:45:36.035307',
u'url': u'/api/histories/a956ab1d3d21799b',
u'user_id': u'bd0631c5dfef81b1',
u'username_and_slug': None}
delete_history()
Delete a history. (Marking it as deleted).
d = gi.histories.delete_history(history_id, purge=False)
if purge=True, also purge (permanently delete) the history (but the Galaxy instance must have the allow_user_dataset_purge option=True in the config/galaxy.ini configuration file.).
There is an apposite method, to mark it as undelete
d = gi.histories.undelete_history(history_id)
update_history()
Update history metadata information.
u = gi.histories.update_history(history_id, **kwds)
Some of the attributes that can be modified are documented below.
- name (str) – Replace history name with the given string
- annotation (str) – Replace history annotation with given string
- deleted (bool) – Mark or unmark history as deleted
- purged (bool) – If True, mark history as purged (permanently deleted). Ignored on Galaxy release_15.01 and earlier
- published (bool) – Mark or unmark history as published
- importable (bool) – Mark or unmark history as importable
- tags (list) – Replace history tags with the given list
Return type: dict
Returns: Dictionary containing details of the updated history
get_status()
Returns the state of this history.
s = gi.histories.get_status(history_id)
Return type: dict
Returns: A dict documenting the current state of the history.
- ‘state’ = This is the current state of the history, such as ok, error, new etc.
- ‘state_details’ = Contains individual statistics for various dataset states.
- ‘percent_complete’ = The overall number of datasets processed to completion.
{'percent_complete': 27,
'state': u'queued',
'state_details': {u'discarded': 0,
u'empty': 0,
u'error': 4,
u'failed_metadata': 0,
u'new': 6,
u'ok': 5,
u'paused': 3,
u'queued': 0,
u'running': 0,
u'setting_metadata': 0,
u'upload': 0}}
Datasets in Histories
show_dataset()
Get details about a given history dataset.
dataset = gi.histories.show_dataset(history_id, dataset_id)
Return type: dict
Returns : Dictionary containing details of the given dataset in the given history
{u'accessible': True,
u'annotation': None,
u'api_type': u'file',
u'create_time': u'2017-01-13T10:29:05.946854',
u'creating_job': u'cf4ef221d2783a89',
u'data_type': u'galaxy.datatypes.tabular.Tabular',
u'dataset_id': u'0674c2fea6a8838a',
u'deleted': True,
u'display_apps': [],
u'display_types': [],
u'download_url': u'/api/histories/7f929922b23b572f/contents/bbd44e69cb8906b5ce7fe31bd9b0016a/display',
u'extension': u'tabular',
u'file_ext': u'tabular',
u'file_size': 974,
u'genome_build': u'?',
u'hda_ldda': u'hda',
u'hid': 1,
u'history_content_type': u'dataset',
u'history_id': u'7f929922b23b572f',
u'id': u'bbd44e69cb8906b5ce7fe31bd9b0016a',
u'meta_files': [],
u'metadata_column_names': None,
u'metadata_column_types': [u'str',
u'float',
u'float',
u'float',
u'float',
u'float',
u'float',
u'float'],
u'metadata_columns': 8,
u'metadata_comment_lines': 1,
u'metadata_data_lines': 20,
u'metadata_dbkey': u'?',
u'metadata_delimiter': u'\t',
u'misc_blurb': u'20 lines, 1 comments',
u'misc_info': u'',
u'model_class': u'HistoryDatasetAssociation',
u'name': u'amino_acid_features.txt',
u'peek': u'<table cellspacing="0" cellpadding="3"><tr><th>1</th><th>2</th><th>3</th><th>4</th><th>5</th><th>6</th><th>7</th><th>8</th></tr><tr><td>#</td><td>Hydrophobicity</td><td>Membrane</td><td>Flexibility</td><td>Volume</td><td>Buried</td><td>Helix</td><td>Beta</td></tr><tr><td>Alanine</td><td>0.61</td><td>1.56</td><td>0.357</td><td>52.6</td><td>91.5</td><td>1.42</td><td>0.83</td></tr><tr><td>Arginine</td><td>0.6</td><td>0.45</td><td>0.529</td><td>109.1</td><td>202</td><td>0.98</td><td>0.93</td></tr><tr><td>Asparagine</td><td>0.06</td><td>0.27</td><td>0.463</td><td>75.7</td><td>135.2</td><td>0.67</td><td>0.89</td></tr><tr><td>Aspartic</td><td>0.46</td><td>0.14</td><td>0.511</td><td>68.4</td><td>124.5</td><td>1.01</td><td>0.54</td></tr><tr><td>Cysteine</td><td>1.07</td><td>1.23</td><td>0.346</td><td>68.3</td><td>117.7</td><td>0.7</td><td>1.19</td></tr></table>',
u'purged': True,
u'rerunnable': False,
u'resubmitted': False,
u'state': u'ok',
u'tags': [],
u'type': u'file',
u'type_id': u'dataset-bbd44e69cb8906b5ce7fe31bd9b0016a',
u'update_time': u'2017-01-28T04:46:48.999395',
u'url': u'/api/histories/7f929922b23b572f/contents/bbd44e69cb8906b5ce7fe31bd9b0016a',
u'uuid': u'93adc5f1-f212-4d31-a84f-a8e39e27821f',
u'visible': True,
u'visualizations': [{u'embeddable': False,
u'href': u'/plugins/visualizations/charts/show?dataset_id=bbd44e69cb8906b5ce7fe31bd9b0016a',
u'html': u'Charts',
u'target': u'galaxy_main'},
{u'embeddable': False,
u'href': u'/plugins/visualizations/scatterplot/show?dataset_id=bbd44e69cb8906b5ce7fe31bd9b0016a',
u'html': u'Scatterplot',
u'target': u'galaxy_main'},
{u'embeddable': False,
u'href': u'/plugins/interactive_environments/jupyter/show?dataset_id=bbd44e69cb8906b5ce7fe31bd9b0016a',
u'html': u'Jupyter',
u'target': u'galaxy_main'}]}
show_dataset_provenance()
Get details related to how dataset was created (id, job_id, tool_id, stdout, stderr, parameters, inputs, etc…).
provenance = gi.histories.show_dataset_provenance(history_id, dataset_id, follow=False)
If follow=True, recursively fetch dataset provenance information for all inputs and their inputs.
Return type: dict
Returns: Dictionary containing provenance of the given dataset in the given history
{u'id': u'cc714f93cb88b900',
u'job_id': u'cf4ef221d2783a89',
u'parameters': {u'async_datasets': u'"None"',
u'dbkey': u'"?"',
u'file_type': u'"auto"',
u'files': u'[{"to_posix_lines": "Yes", "NAME": "None", "file_data": "/galaxy-repl/main/upload/0018550056", "space_to_tab": null, "url_paste": "None", "__index__": 0, "ftp_files": null, "uuid": "None"}]',
u'files_metadata': u'{"file_type": "auto", "__current_case__": 31}',
u'paramfile': u'"/galaxy/main/scratch/tmp_I9NFp"'},
u'stderr': u'',
u'stdout': u'',
u'tool_id': u'upload1',
u'uuid': u'93adc5f1-f212-4d31-a84f-a8e39e27821f'}
update_dataset()
Update history dataset metadata.
u = gi.histories.update_dataset(history_id, dataset_id, **kwds)
Some of the attributes that can be modified are documented below.
- name (str) – Replace history dataset name with the given string
- genome_build (str) – Replace history dataset genome build (dbkey)
- annotation (str) – Replace history dataset annotation with given string
- deleted (bool) – Mark or unmark history dataset as deleted
- visible (bool) – Mark or unmark history dataset as visible
Return type: dict
Returns: Dictionary containing details of the updated dataset.
delete_dataset()
Mark corresponding dataset as deleted.
d = gi.histories.delete_dataset(history_id, dataset_id, purge=False)
if purge=True, also purge (permanently delete) the dataset. (but the Galaxy instance must have the allow_user_dataset_purge=True in the config/galaxy.ini configuration file.).
upload_dataset_from_library()
Upload a dataset into the history from a library.
u = gi.histories.upload_dataset_from_library(history_id, lib_dataset_id)
Requires the library dataset ID, which can be obtained from the library contents.
Examples
Workflows
Contains possible interactions with the Galaxy Workflows (View source)
class bioblend.galaxy.workflows.WorkflowClient(galaxy_instance)
get_workflows()
Get all workflows or filter the specific one(s) via the provided name or workflow_id.
workflows = gi.workflows.get_workflows(workflow_id=None, name=None, published=False)
Provide only one argument, name or workflow_id, but not both.
if published=True, return also published workflows
Return type: list
Returns: A list of workflow dicts
[{u'deleted': False,
u'id': u'387220538943a562',
u'latest_workflow_uuid': u'48b0f7e1-3f5f-4e84-bad6-d8ca86b085f2',
u'model_class': u'StoredWorkflow',
u'name': u"imported: 'Galaxy 101'",
u'owner': u'ratzeni',
u'published': False,
u'tags': [],
u'url': u'/api/workflows/387220538943a562'}]
show_workflow()
Display information needed to run a workflow.
workflow = gi.worflows.show_workflow(workflow_id)
Return type: dict
Returns: A description of the workflow and its inputs.
{u'id': u'92c56938c2f9b315',
u'inputs': {u'23': {u'label': u'Input Dataset', u'value': u''}},
u'name': u'Simple',
u'url': u'/api/workflows/92c56938c2f9b315'}
{u'annotation': None,
u'deleted': False,
u'id': u'387220538943a562',
u'inputs': {u'0': {u'label': u'Input Dataset',
u'uuid': u'999d5b6e-75fa-4f28-81c8-0ceab17c7b1b',
u'value': u''},
u'1': {u'label': u'Input Dataset',
u'uuid': u'beee39e4-469e-400d-8621-20c824670fdc',
u'value': u''}},
u'latest_workflow_uuid': u'48b0f7e1-3f5f-4e84-bad6-d8ca86b085f2',
u'model_class': u'StoredWorkflow',
u'name': u"imported: 'Galaxy 101'",
u'owner': u'ratzeni',
u'published': False,
u'steps': {u'0': {u'annotation': None,
u'id': 0,
u'input_steps': {},
u'tool_id': None,
u'tool_inputs': {u'name': u'Input Dataset'},
u'tool_version': None,
u'type': u'data_input'},
u'1': {u'annotation': None,
u'id': 1,
u'input_steps': {},
u'tool_id': None,
u'tool_inputs': {u'name': u'Input Dataset'},
u'tool_version': None,
u'type': u'data_input'},
u'2': {u'annotation': None,
u'id': 2,
u'input_steps': {u'input1': {u'source_step': 0, u'step_output': u'output'},
u'input2': {u'source_step': 1, u'step_output': u'output'}},
u'tool_id': u'toolshed.g2.bx.psu.edu/repos/devteam/join/gops_join_1/1.0.0',
u'tool_inputs': {u'chromInfo': u'"/galaxy-repl/localdata/chrom/hg19.len"',
u'fill': u'"none"',
u'input1': u'null',
u'input2': u'null',
u'min': u'"1"'},
u'tool_version': None,
u'type': u'tool'},
u'3': {u'annotation': None,
u'id': 3,
u'input_steps': {u'input1': {u'source_step': 2, u'step_output': u'output'}},
u'tool_id': u'Grouping1',
u'tool_inputs': {u'chromInfo': u'"/galaxy-repl/localdata/chrom/hg19.len"',
u'groupcol': u'{"__class__": "UnvalidatedValue", "value": "4"}',
u'ignorecase': u'"False"',
u'ignorelines': u'null',
u'input1': u'null',
u'operations': u'[{"opcol": {"__class__": "UnvalidatedValue", "value": "4"}, "__index__": 0, "optype": "length", "opround": "no"}]'},
u'tool_version': None,
u'type': u'tool'},
u'4': {u'annotation': None,
u'id': 4,
u'input_steps': {u'input': {u'source_step': 3,
u'step_output': u'out_file1'}},
u'tool_id': u'sort1',
u'tool_inputs': {u'chromInfo': u'"/galaxy-repl/localdata/chrom/hg19.len"',
u'column': u'{"__class__": "UnvalidatedValue", "value": "2"}',
u'column_set': u'[]',
u'input': u'null',
u'order': u'"DESC"',
u'style': u'"num"'},
u'tool_version': None,
u'type': u'tool'},
u'5': {u'annotation': None,
u'id': 5,
u'input_steps': {u'input': {u'source_step': 4,
u'step_output': u'out_file1'}},
u'tool_id': u'Show beginning1',
u'tool_inputs': {u'chromInfo': u'"/galaxy-repl/localdata/chrom/hg19.len"',
u'input': u'null',
u'lineNum': u'"5"'},
u'tool_version': None,
u'type': u'tool'},
u'6': {u'annotation': None,
u'id': 6,
u'input_steps': {u'input1': {u'source_step': 0, u'step_output': u'output'},
u'input2': {u'source_step': 5, u'step_output': u'out_file1'}},
u'tool_id': u'comp1',
u'tool_inputs': {u'chromInfo': u'"/galaxy-repl/localdata/chrom/hg19.len"',
u'field1': u'{"__class__": "UnvalidatedValue", "value": "4"}',
u'field2': u'{"__class__": "UnvalidatedValue", "value": "1"}',
u'input1': u'null',
u'input2': u'null',
u'mode': u'"N"'},
u'tool_version': None,
u'type': u'tool'}},
u'tags': [],
u'url': u'/api/workflows/387220538943a562'}
Import/Export
export_workflow_dict()
Exports a workflow.
workflow = gi.workflows.export_workflow_dict(workflow_id)
Return type: dict
Returns: Dictionary representing the requested workflow
{u'a_galaxy_workflow': u'true',
u'annotation': u'',
u'format-version': u'0.1',
u'name': u" 'Galaxy 101'",
u'steps': {u'0': {u'annotation': u'',
u'content_id': None,
u'errors': None,
u'id': 0,
u'input_connections': {},
u'inputs': [{u'description': u'', u'name': u'Input Dataset'}],
u'label': None,
u'name': u'Input dataset',
u'outputs': [],
u'position': {u'left': 10, u'top': 10},
u'tool_id': None,
u'tool_state': u'{"name": "Input Dataset"}',
u'tool_version': None,
u'type': u'data_input',
u'uuid': u'999d5b6e-75fa-4f28-81c8-0ceab17c7b1b',
u'workflow_outputs': []},
u'1': {u'annotation': u'',
u'content_id': None,
u'errors': None,
u'id': 1,
u'input_connections': {},
u'inputs': [{u'description': u'', u'name': u'Input Dataset'}],
u'label': None,
u'name': u'Input dataset',
u'outputs': [],
u'position': {u'left': 10, u'top': 130},
u'tool_id': None,
u'tool_state': u'{"name": "Input Dataset"}',
u'tool_version': None,
u'type': u'data_input',
u'uuid': u'beee39e4-469e-400d-8621-20c824670fdc',
u'workflow_outputs': []},
u'2': {u'annotation': u'',
u'content_id': u'toolshed.g2.bx.psu.edu/repos/devteam/join/gops_join_1/1.0.0',
u'errors': None,
u'id': 2,
u'input_connections': {u'input1': {u'id': 0, u'output_name': u'output'},
u'input2': {u'id': 1, u'output_name': u'output'}},
u'inputs': [],
u'label': None,
u'name': u'Join',
u'outputs': [{u'name': u'output', u'type': u'interval'}],
u'position': {u'left': 230, u'top': 10},
u'post_job_actions': {},
u'tool_id': u'toolshed.g2.bx.psu.edu/repos/devteam/join/gops_join_1/1.0.0',
u'tool_shed_repository': {u'changeset_revision': u'de21bdbb8d28',
u'name': u'join',
u'owner': u'devteam',
u'tool_shed': u'toolshed.g2.bx.psu.edu'},
u'tool_state': u'{"input2": "null", "__page__": null, "input1": "null", "min": "\\"1\\"", "__rerun_remap_job_id__": null, "chromInfo": "\\"/galaxy-repl/localdata/chrom/hg19.len\\"", "fill": "\\"none\\""}',
u'tool_version': None,
u'type': u'tool',
u'uuid': u'6cdd56e0-b42c-4fac-a2d3-f09965181fd1',
u'workflow_outputs': []},
u'3': {u'annotation': u'',
u'content_id': u'Grouping1',
u'errors': None,
u'id': 3,
u'input_connections': {u'input1': {u'id': 2, u'output_name': u'output'}},
u'inputs': [],
u'label': None,
u'name': u'Group',
u'outputs': [{u'name': u'out_file1', u'type': u'tabular'}],
u'position': {u'left': 450, u'top': 10},
u'post_job_actions': {},
u'tool_id': u'Grouping1',
u'tool_state': u'{"operations": "[{\\"opcol\\": \\"4\\", \\"__index__\\": 0, \\"optype\\": \\"length\\", \\"opround\\": \\"no\\"}]", "__page__": null, "input1": "null", "ignorelines": "null", "groupcol": "\\"4\\"", "__rerun_remap_job_id__": null, "ignorecase": "\\"false\\"", "chromInfo": "\\"/galaxy-repl/localdata/chrom/hg19.len\\""}',
u'tool_version': None,
u'type': u'tool',
u'uuid': u'6a242308-bf32-4776-b54c-6499198a9a8b',
u'workflow_outputs': []},
u'4': {u'annotation': u'',
u'content_id': u'sort1',
u'errors': None,
u'id': 4,
u'input_connections': {u'input': {u'id': 3, u'output_name': u'out_file1'}},
u'inputs': [],
u'label': None,
u'name': u'Sort',
u'outputs': [{u'name': u'out_file1', u'type': u'input'}],
u'position': {u'left': 670, u'top': 10},
u'post_job_actions': {},
u'tool_id': u'sort1',
u'tool_state': u'{"__page__": null, "style": "\\"num\\"", "column": "\\"2\\"", "__rerun_remap_job_id__": null, "order": "\\"DESC\\"", "input": "null", "chromInfo": "\\"/galaxy-repl/localdata/chrom/hg19.len\\"", "column_set": "[]"}',
u'tool_version': None,
u'type': u'tool',
u'uuid': u'07002a67-898e-4a96-9566-72e0d8fb684a',
u'workflow_outputs': []},
u'5': {u'annotation': u'',
u'content_id': u'Show beginning1',
u'errors': None,
u'id': 5,
u'input_connections': {u'input': {u'id': 4, u'output_name': u'out_file1'}},
u'inputs': [],
u'label': None,
u'name': u'Select first',
u'outputs': [{u'name': u'out_file1', u'type': u'input'}],
u'position': {u'left': 890, u'top': 10},
u'post_job_actions': {},
u'tool_id': u'Show beginning1',
u'tool_state': u'{"__page__": null, "input": "null", "__rerun_remap_job_id__": null, "chromInfo": "\\"/galaxy-repl/localdata/chrom/hg19.len\\"", "lineNum": "\\"5\\""}',
u'tool_version': None,
u'type': u'tool',
u'uuid': u'56462532-69b6-4e22-b01e-ff3656a552ba',
u'workflow_outputs': []},
u'6': {u'annotation': u'',
u'content_id': u'comp1',
u'errors': None,
u'id': 6,
u'input_connections': {u'input1': {u'id': 0, u'output_name': u'output'},
u'input2': {u'id': 5, u'output_name': u'out_file1'}},
u'inputs': [],
u'label': None,
u'name': u'Compare two Datasets',
u'outputs': [{u'name': u'out_file1', u'type': u'input'}],
u'position': {u'left': 1110, u'top': 10},
u'post_job_actions': {},
u'tool_id': u'comp1',
u'tool_state': u'{"input2": "null", "__page__": null, "input1": "null", "field2": "\\"1\\"", "__rerun_remap_job_id__": null, "field1": "\\"4\\"", "mode": "\\"N\\"", "chromInfo": "\\"/galaxy-repl/localdata/chrom/hg19.len\\""}',
u'tool_version': None,
u'type': u'tool',
u'uuid': u'3b17ef01-db64-429b-ae49-6d1fcd610da2',
u'workflow_outputs': []}},
u'uuid': u'48b0f7e1-3f5f-4e84-bad6-d8ca86b085f2'}
export_workflow_to_local_path()
Exports a workflow in JSON format to a given local path.
gi.workflows.export_workflow_to_local_path(workflow_id, file_local_path, use_default_filename=True)
file_local_path(str) – Local path to which the exported file will be saved. (Should not contain filename ifuse_default_name=True)use_default_filename(bool) – IfTrue, the exported file will be saved asfile_local_path/Galaxy-Workflow-%s.ga,where %s is the workflow name.
import_workflow_dict()
Imports a new workflow given a dictionary representing a previously exported workflow.
gi.workflows.import_workflow_dict(workflow_dict)
import_workflow_from_local_path()
Imports a new workflow given the path to a file containing a previously exported workflow.
gi.workflows.import_workflow_from_local_path(file_local_path)
import_shared_workflow()
Imports a new workflow from the shared published workflows.
gi.workflows.import_shared_workflow(workflow_id)
Return type: dict
Returns: A description of the workflow.
get_workflow_inputs()
Get a list of workflow input IDs that match the given label.
inputs = gi.workflows.get_workflow_inputs(workflow_id, label)
If no input matches the given label, an empty list is returned.
[u'1', u'0']
invoke_workflow()
Run the workflow identified by workflow_id.
This will cause a workflow to be scheduled and return an object describing the workflow invocation.
w = gi.workflows.invoke_workflow(workflow_id, inputs=None, params=None, history_id=None, history_name=None, import_inputs_to_history=False, replacement_params=None, allow_tool_state_corrections=None)
Parameters:
workflow_id(str) – Encoded workflow IDinputs(dict) – A mapping of workflow inputs to datasets and dataset collections.
The datasets source can be a LibraryDatasetDatasetAssociation (ldda), LibraryDataset (ld), HistoryDatasetAssociation (hda), or HistoryDatasetCollectionAssociation (hdca).
The map must be in the following format:
{'<input_index>': {'id': <encoded dataset ID>, 'src': '[ldda, ld, hda, hdca]'}}(e.g.{'2': {'id': '29beef4fadeed09f', 'src': 'hda'}})
This map may also be indexed by the UUIDs of the workflow steps, as indicated by theuuidproperty of steps returned from the Galaxy API.params(dict) – A mapping of non-datasets tool parameters (see below)history_id(str) – The encoded history ID where to store the workflow output. Alternatively,history_name maybe specified to create a new history.history_name(str) – Create a new history with the given name to store the workflow output. If bothhistory_idandhistory_nameare provided,history_nameis ignored. If neither is specified, a newUnnamed historyis created.import_inputs_to_history(bool) – IfTrue, used workflow inputs will be imported into the history. IfFalse, only workflow outputs will be visible in the given history.allow_tool_state_corrections(bool) – IfTrue, allow Galaxy to fill in missing tool state when running workflows. This may be useful for workflows using tools that have changed over time or for workflows built outside of Galaxy with only a subset of inputs defined.replacement_params(dict) – pattern-based replacements for post-job actions (see below)
The params dict should be specified as follows:
{STEP_ID: PARAM_DICT, ...}
where PARAM_DICT is:
{PARAM_NAME: VALUE, ...}
The replacement_params dict should map parameter names in post-job actions (PJAs) to their runtime values.
For instance, if the final step has a PJA like the following:
{u'RenameDatasetActionout_file1':
{u'action_arguments': {u'newname': u'${output}'},
u'action_type': u'RenameDatasetAction',
u'output_name': u'out_file1'
}
}
then the following renames the output dataset to ‘foo’:
replacement_params = {'output': 'foo'}
Return type: dict
Returns: A dict containing the workflow invocation describing the scheduling of the workflow.
u'history_id': u'2f94e8ae9edff68a',
u'id': u'df7a1f0c02a5b08e',
u'inputs': {u'0': {u'id': u'a7db2fac67043c7e',
u'src': u'hda',
u'uuid': u'7932ffe0-2340-4952-8857-dbaa50f1f46a'}},
u'model_class': u'WorkflowInvocation',
u'state': u'ready',
u'steps': [{u'action': None,
u'id': u'd413a19dec13d11e',
u'job_id': None,
u'model_class': u'WorkflowInvocationStep',
u'order_index': 0,
u'state': None,
u'update_time': u'2015-10-31T22:00:26',
u'workflow_step_id': u'cbbbf59e8f08c98c',
u'workflow_step_label': None,
u'workflow_step_uuid': u'b81250fd-3278-4e6a-b269-56a1f01ef485'},
{u'action': None,
u'id': u'2f94e8ae9edff68a',
u'job_id': u'e89067bb68bee7a0',
u'model_class': u'WorkflowInvocationStep',
u'order_index': 1,
u'state': u'new',
u'update_time': u'2015-10-31T22:00:26',
u'workflow_step_id': u'964b37715ec9bd22',
u'workflow_step_label': None,
u'workflow_step_uuid': u'e62440b8-e911-408b-b124-e05435d3125e'}],
u'update_time': u'2015-10-31T22:00:26',
u'uuid': u'c8aa2b1c-801a-11e5-a9e5-8ca98228593c',
u'workflow_id': u'03501d7626bd192f'}
run_workflow()
Run the workflow identified by workflow_id. Deprecated since version 0.7.0: Use invoke_workflow() instead.
gi.workflows.run_workflow(workflow_id, dataset_map=None, params=None, history_id=None, history_name=None, import_inputs_to_history=False, replacement_params=None)
Historically, the run_workflow method consumed a dataset_map data structure that was indexed by unencoded workflow step IDs.
These IDs would not be stable across Galaxy instances. The new inputs property of invoke_workflow is instead indexed
by either the order_index property (which is stable across workflow imports) or the step UUID which is also stable.
Examples
Tools
Contains possible interaction dealing with Galaxy tools. (View source)
class bioblend.galaxy.tools.ToolClient(galaxy_instance)
get_tools()
Get all tools or filter the specific one(s) via the provided name or tool_id.
tools = gi.tools.get_tools(tool_id=None, name=None, trackster=None)
Provide only one argument, name or tool_id, but not both.
If name is set and multiple names match the given name, all the tools matching the argument will be returned.
if trackster=True, only tools that are compatible with Trackster are returned
Return type: list
Returns: List of tool descriptions.
[{u'description': u'table browser',
u'edam_operations': [],
u'edam_topics': [],
u'form_style': u'special',
u'id': u'ucsc_table_direct1',
u'labels': [],
u'link': u'/tool_runner/data_source_redirect?tool_id=ucsc_table_direct1',
u'min_width': u'800',
u'model_class': u'DataSourceTool',
u'name': u'UCSC Main',
u'panel_section_id': u'getext',
u'panel_section_name': u'Get Data',
u'target': u'_top',
u'version': u'1.0.0'},
{u'description': u'Get otus for each distance in a otu list',
u'edam_operations': [],
u'edam_topics': [],
u'form_style': u'regular',
u'id': u'toolshed.g2.bx.psu.edu/repos/jjohnson/mothur_toolsuite/mothur_get_otulist/1.20.0',
u'labels': [],
u'link': u'/tool_runner?tool_id=toolshed.g2.bx.psu.edu%2Frepos%2Fjjohnson%2Fmothur_toolsuite%2Fmothur_get_otulist%2F1.20.0',
u'min_width': -1,
u'model_class': u'Tool',
u'name': u'Get.otulist',
u'panel_section_id': u'ngs:_mothur',
u'panel_section_name': u'NGS: Mothur',
u'target': u'galaxy_main',
u'tool_shed_repository': {u'changeset_revision': u'040410b8167e',
u'name': u'mothur_toolsuite',
u'owner': u'jjohnson',
u'tool_shed': u'toolshed.g2.bx.psu.edu'},
u'version': u'1.20.0'},]
show_tool()
Get details of a given tool.
tool = gi.tools.show_tool(tool_id, io_details=False, link_details=False)
When io_details=True, get also input and output details
When link_details=True, get also link details
Return type: dict
Returns: A description of the given tool
{u'description': u'table browser',
u'edam_operations': [],
u'edam_topics': [],
u'form_style': u'special',
u'id': u'ucsc_table_direct1',
u'labels': [],
u'model_class': u'DataSourceTool',
u'name': u'UCSC Main',
u'panel_section_id': u'getext',
u'panel_section_name': u'Get Data',
u'version': u'1.0.0'}
upload_file()
Upload the file specified by path to the history specified by history_id.
gi.tools.upload_file(path, history_id, **keywords)
file_name(str) – (optional) name of the new history datasetfile_type(str) – Galaxy datatype for the new dataset, default is autodbkey(str) – (optional) genome dbkeyto_posix_lines(bool) – ifTrue, convert universal line endings to POSIX line endings. Default isTrue. Set to False if you upload a gzip, bz2 or zip archive containing a binary filespace_to_tab(bool) – whether to convert spaces to tabs. Default is False. Applicable only if to_posix_lines is True
run_tool()
Runs tool specified by tool_id in history indicated by history_id with inputs from dict tool_inputs.
gi.tools.run_tool(history_id, tool_id, tool_inputs)
The tool_inputs dict should contain input datasets and parameters in the (largely undocumented) format used by the Galaxy API.
Some examples can be found here
Jobs
Contains possible interactions with the Galaxy Jobs (View source)
class bioblend.galaxy.jobs.JobsClient(galaxy_instance)
get_jobs()
Get the list of jobs of the current user.
jobs = gi.jobs.get_jobs()
Return type: list
Returns: list of dictionaries containing summary job information.
[{u'create_time': u'2017-01-13T14:30:34.849014',
u'exit_code': None,
u'id': u'bbd44e69cb8906b5c20764c7df14bca4',
u'model_class': u'Job',
u'state': u'error',
u'tool_id': u'Grouping1',
u'update_time': u'2017-01-13T14:30:46.647542'},
{u'create_time': u'2017-01-13T14:30:35.278443',
u'exit_code': None,
u'id': u'bbd44e69cb8906b5ae0ed21eed93eef1',
u'model_class': u'Job',
u'state': u'paused',
u'tool_id': u'sort1',
u'update_time': u'2017-01-13T14:30:46.604752'},
{u'create_time': u'2017-01-13T14:30:34.222427',
u'exit_code': 0,
u'id': u'bbd44e69cb8906b5dc07e598c63ef87c',
u'model_class': u'Job',
u'state': u'ok',
u'tool_id': u'toolshed.g2.bx.psu.edu/repos/devteam/join/gops_join_1/1.0.0',
u'update_time': u'2017-01-13T14:30:45.337764'},]
show_job()
Get details of a given job of the current user.
job = gi.jobs.show_job(job_id, full_details=False)
When full_details=True,it shows the complete list of details for the given job.
Return type: dict
Returns: A description of the given job
{u'create_time': u'2017-01-13T14:30:34.849014',
u'exit_code': None,
u'id': u'bbd44e69cb8906b5c20764c7df14bca4',
u'inputs': {u'input1': {u'id': u'bbd44e69cb8906b5b11e71f4d1889ae2',
u'src': u'hda',
u'uuid': u'd501eb5d-f9cd-4fcc-b2aa-1484428f0c99'}},
u'model_class': u'Job',
u'outputs': {u'out_file1': {u'id': u'bbd44e69cb8906b54ce551cbcb48857f',
u'src': u'hda',
u'uuid': u'3316f5e9-1a16-4cc1-b115-20a8de482c04'}},
u'params': {u'__workflow_invocation_uuid__': u'"d5fb4116d99c11e6920e005056a52a46"',
u'chromInfo': u'"/galaxy-repl/localdata/chrom/?.len"',
u'dbkey': u'"?"',
u'groupcol': u'"4"',
u'ignorecase': u'"false"',
u'ignorelines': u'null',
u'operations': u'[{"opcol": "4", "__index__": 0, "optype": "length", "opround": "no"}]'},
u'state': u'error',
u'tool_id': u'Grouping1',
u'update_time': u'2017-01-13T14:30:46.647542'}
get_state()
Display the current state for a given job of the current user.
state = gi.jobs.get_state(job_id)
Return type: str Returns: state of the given job among the following values. If the state cannot be retrieved, an empty string is returned
newqueuedrunningwaitingokerror
Hands On
Go to a directory of you choice and clone this repository:
$ git clone https://github.com/ratzeni/bioblend-tutorial.git
Cd into the tutorial directory
$ cd bioblend-tutorial
Create a new virtual environment in folder .venv
$ virtualenv .venv
virtualenv is a tool to create isolated Python environments. virtualenv creates a folder which contains all the necessary executables to use the packages that a Python project would need.
To begin using the virtual environment, it needs to be activated:
$ source .venv/bin/activate
The name of the current virtual environment will now appear on the left of the prompt to let you know that it’s active.
From now on, any package that you install using pip will be placed in the my_project folder, isolated from the global Python installation.
 Now, install bioblend
Now, install bioblend
$ pip install bioblend
Install jupyter
$ pip install jupyter
Congratulations, you have installed Jupyter Notebook. To run the notebook:
$ jupyter notebook
The Jupyter Notebook is an open-source web application that allows you to create and share documents that contain live code, equations, visualizations and explanatory text. Uses include: data cleaning and transformation, numerical simulation, statistical modeling, machine learning and much more.
Installing with Conda
$ conda create -n bvenv bioblend jupyter
Exiting
Ctrl-c to exit from notebook.
When ou are done working in the virtual environment, you can deactivate it:
$ deactivate