Why Galaxy Tools
Join the Galaxy Developers community and develop your own tools
Why Galaxy tools
- Galaxy Tool Shed
- Tool wrappers, dependencies
- Install and configure your own Galaxy
Tools
- Each tool is a text file describing:
- the input datasets and their datatypes
- the tool parameters (numerical, text, boolean, selections)
- how to generate a command to execute the tool with the specified inputs and parameters
- the output datasets the tool should produce and their datatypes
- help, tests, citations, dependency requirements
Brief history of Galaxy tools
- Local tools only. Admin manually adds tools to
tool_conf.xmland installs the dependencies.- Low reproducibility. Admin overhead
- Galaxy packages. Admin manually creates
env.shfile at the given dependency path that, once sourced, provides the proper binaries.- Better reproducibility. Admin overhead
- Tool Shed packages. Admins let Galaxy install dependencies based on TS ‘recipes’.
- Galaxy-only solution.
- Building another package manager…
- Embracing Conda package manager.
- Galaxy can already resolve dependencies using Conda.
- In close future Conda will be auto-init during Galaxy startup.
- Many prime tools already support it
What is a Tool Shed?

-
Tool Shed is a free tool store, with thousands of tools already available
- For Galaxy administrators (only admin can install tools), facilitates:
- installing/updating tools
- For tool developers, facilitates:
- sharing of Galaxy utilities
- versioning: multiple installable revisions of any repository can be present in Galaxy
Tool Shed Vocabulary
wrapperortool definition file- The XML file that describes to Galaxy how the underlying software works, allowing Galaxy to render UI and execute the software in the right wayrepository- A versioned code archive with tool(s) in Tool Shed
Available Tool Sheds
- Galaxy community Tool Shed - Main Tool Shed
- Galaxy community test Tool Shed - Sandbox for testing
- Other public Tool Sheds
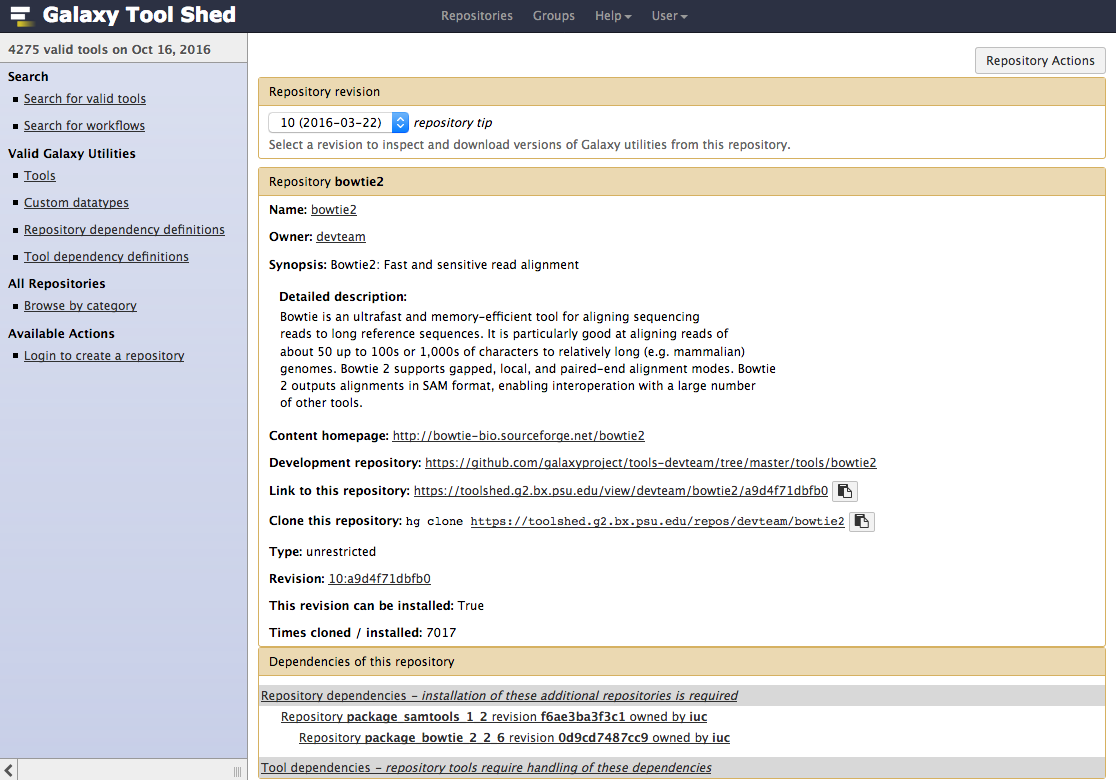
Tool Shed interface
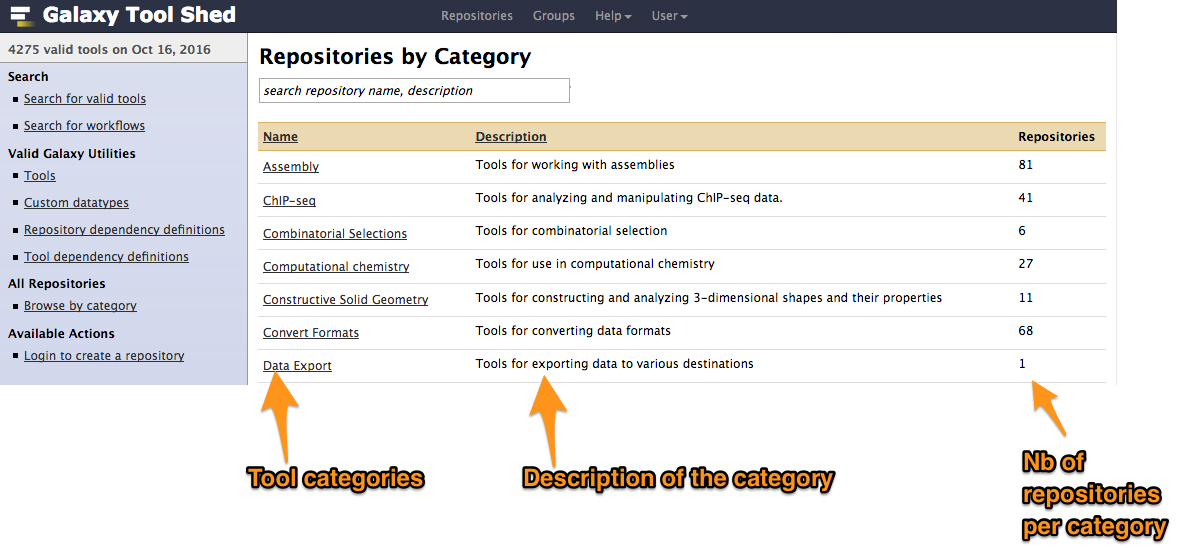
Example of tool
__
Installing tools from a Tool Shed
Connect your Galaxy to a Tool Shed
- Only for third party toolsheds (Main and Test by default)
- In
config/tool_sheds_conf.xml:<tool_sheds> <tool_shed name="Galaxy main tool shed" url="https://toolshed.g2.bx.psu.edu/" /> </tool_sheds> - Restart Galaxy
Install a tool from the Tool Shed
G- o to the admin interface and click on “Search Tool Shed”
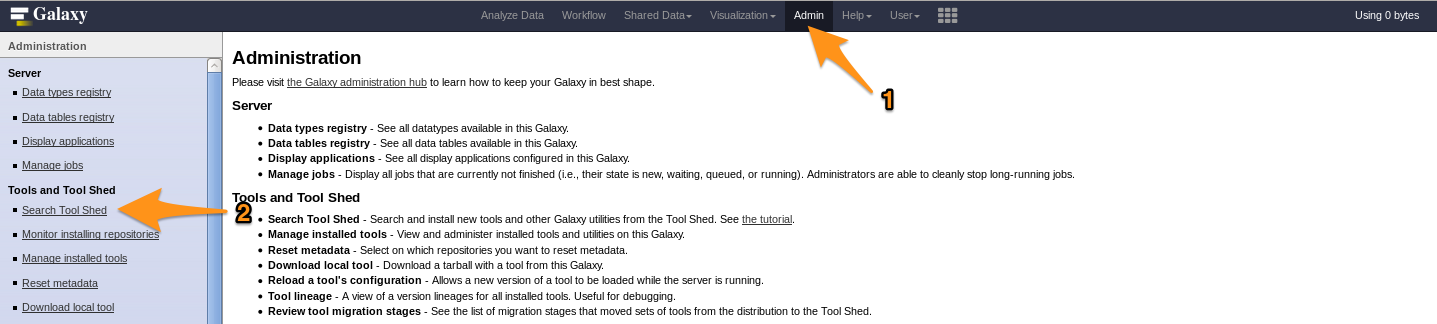
-
Select a Tool Shed
-
Search your tool
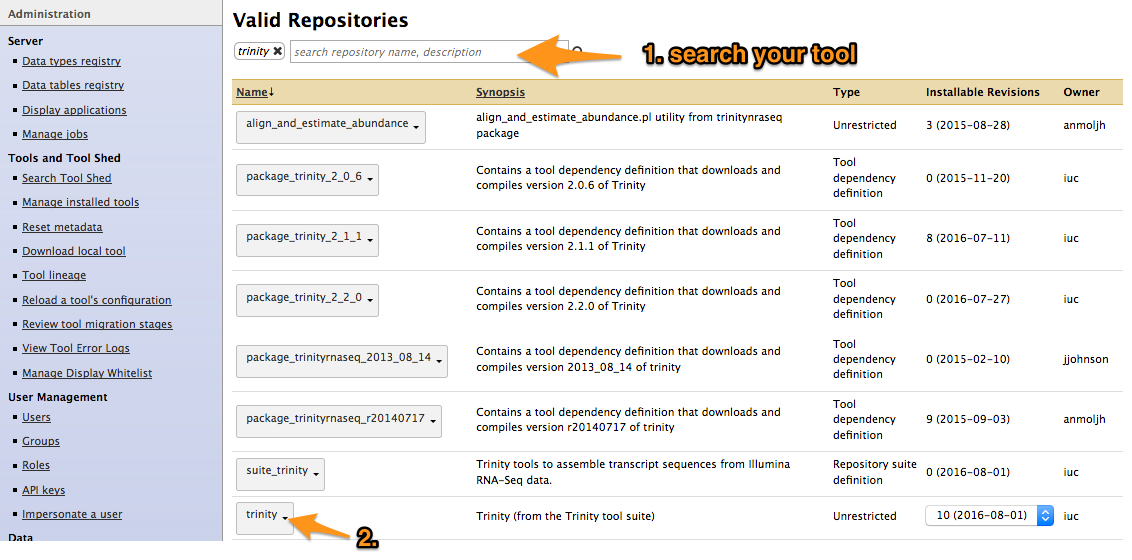
-
Types of repository
- Tools (
$name) - Tool suites (
suite_$name) - Tool dependencies (
package_$name_$version) (gradually removed, replaced by Conda dependencies)
- Tools (
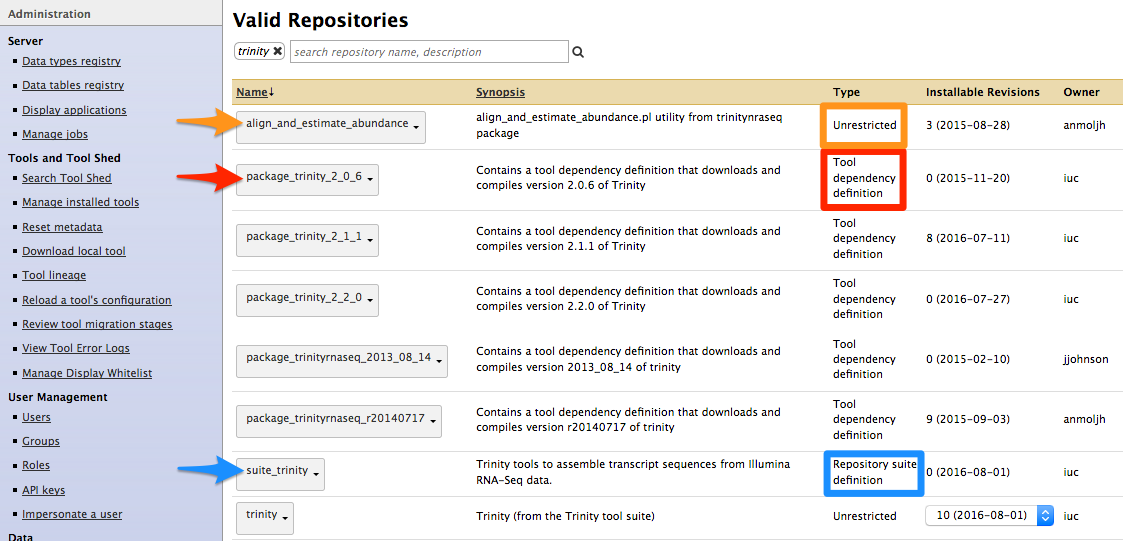
-
Install the tool
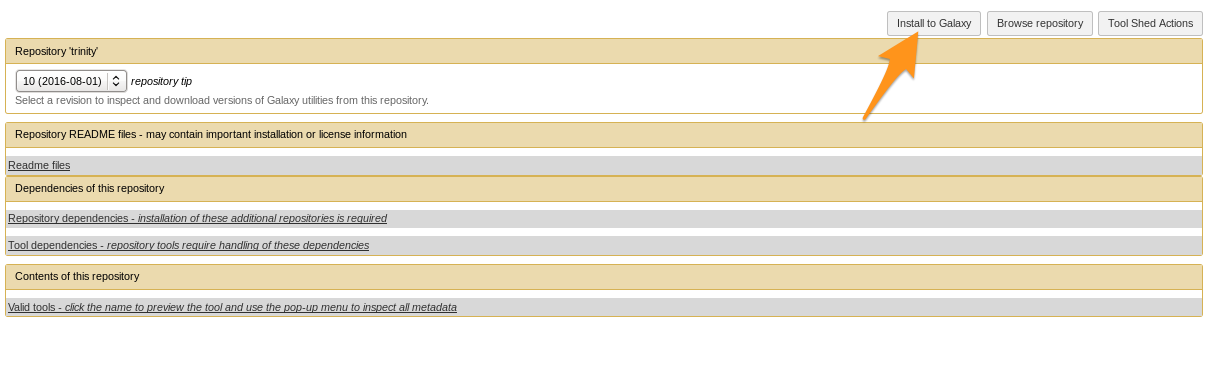
- Install the tool
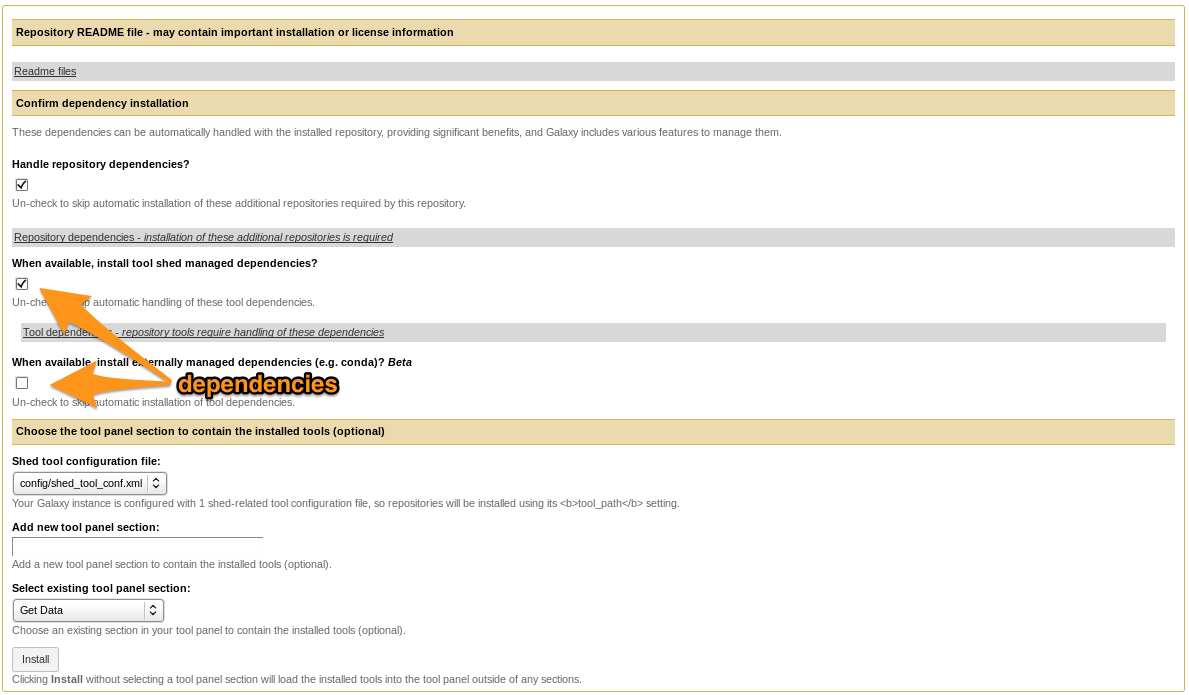
- Selecting Tool Shed AND conda will make Galaxy to install both
-
Recommended: use conda
- Check
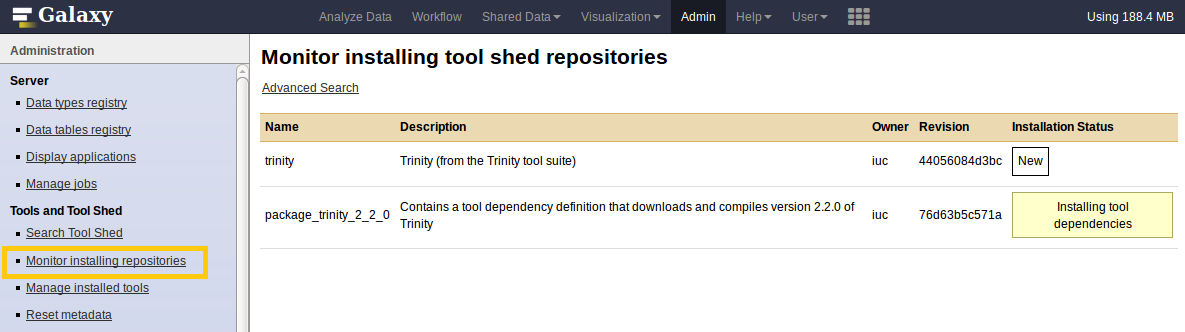
What happened?
- Repository was downloaded.
- If needed Galaxy downloaded and compiled the needed dependencies.
- Galaxy created an entry for the tool in the DB.
- Galaxy added the tool to one of the tool configs (
shed_tool_conf.xml). - After restart Galaxy will load the tool.
Manage installed tools
- Admin - Manage installed tools
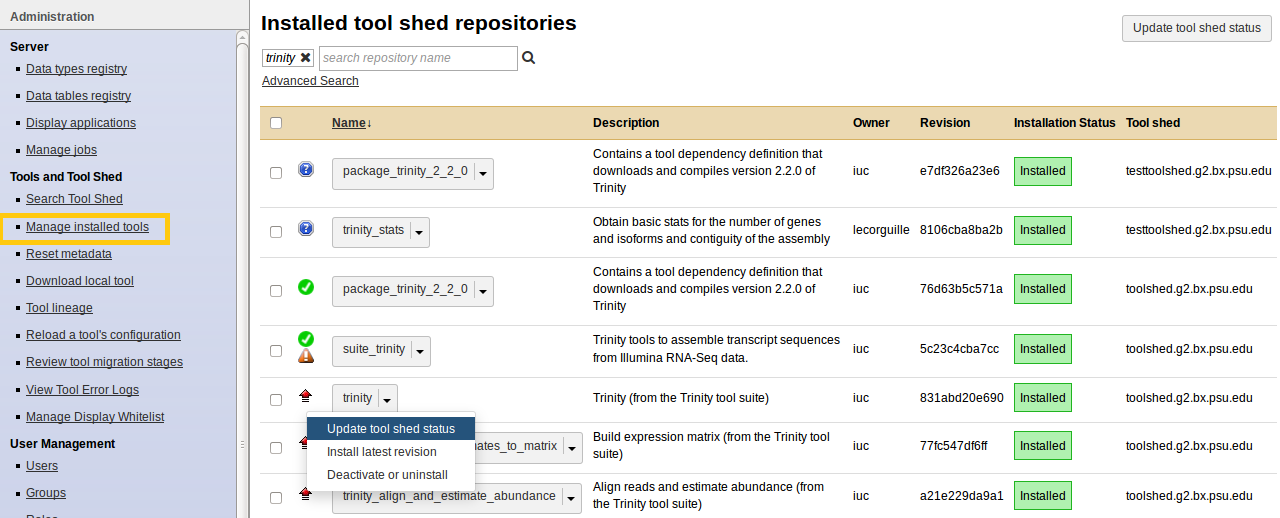

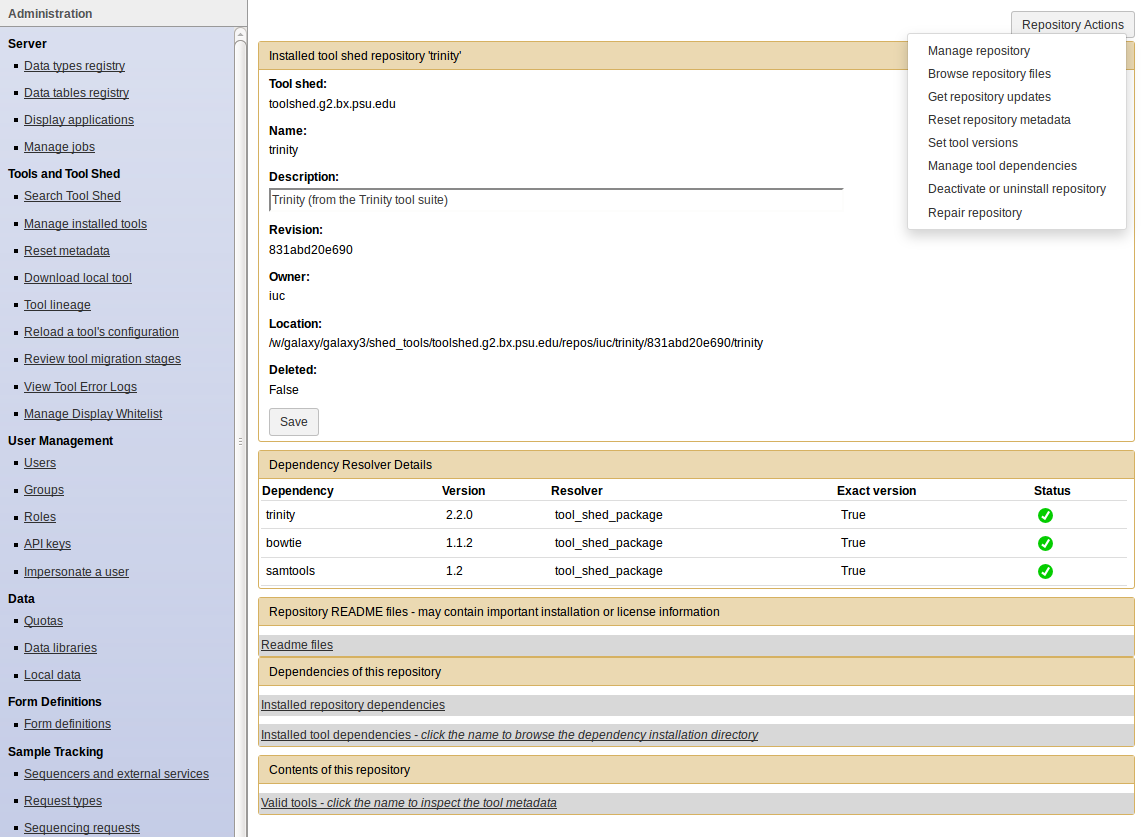
- Click on the name of a tool
- Manage and browse the repository
Tool suite
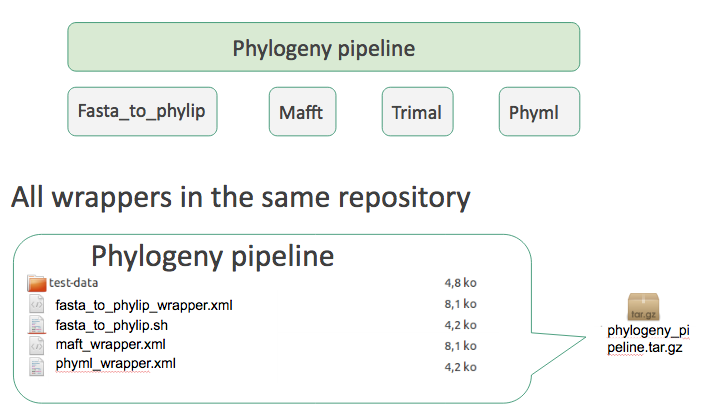
- To install multiple repositories some authors offer suites.
- Suite is a single repository that ‘depends’ on many other.
-
If you install the suite all ‘dependency repositories’ will be installed too.
- One repository per individual tool
- A special ‘Tool suite’ repository with one XML file listing the individual repositories
<?xml version="1.0"?>
<repositories description="Pipeline phylogeny">
<repository toolshed="http://testtoolshed.g2.bx.psu.edu"
name="fasta_to_phylip" owner="gandres" changeset_revision="a895633568" />
<repository name="mafft" owner="gandres" />
<repository name="phyml" owner="gandres" />
[…]
</repositories>
Contributing to a community
Many tools developed by the community on GitHub repositories
Added value:
- Easier development for developers
- Easier contribution for user
- Automated tests on each contribution
- Automated publishing to ToolShed
Galaxy Tool Dependencies
To achieve the level of reproducibility Galaxy aims for, it needs to be able to:
- Install any tool at any version with the exact same dependencies at any time.
Linux/MacOS package management is/was:
- missing the scientific packages
- avoiding or not maintaining old versions
Dependency resolution
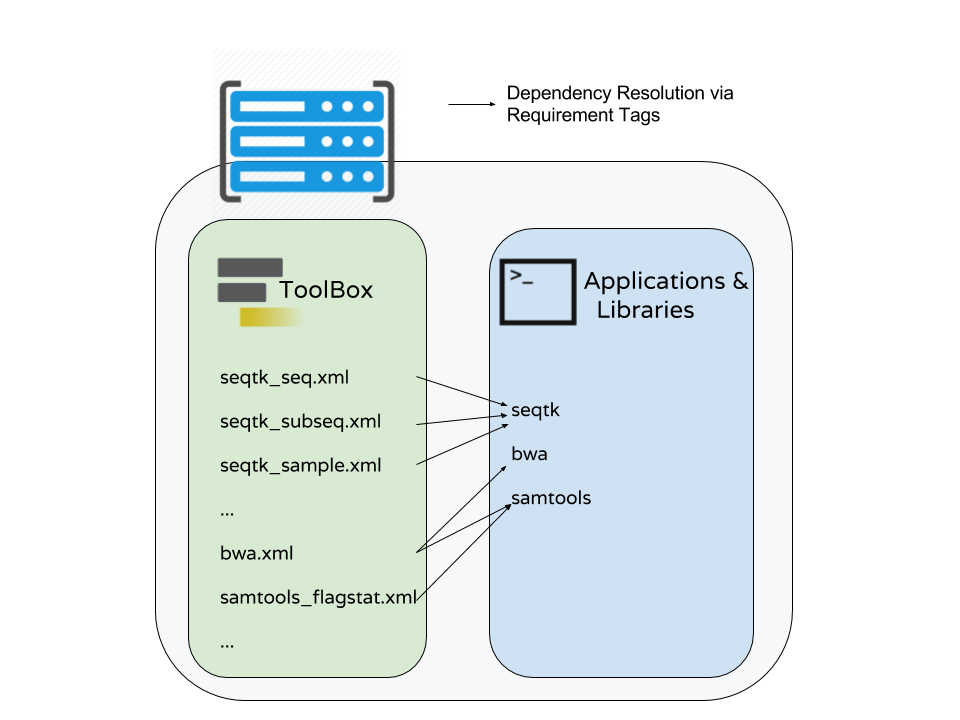
- Multiple tools may be mapped to the same requirements
- There are few different ways to populate Applications and Libraries on the right - we will talk about Conda which is what we consider the “community best practice”.
Conda Key Features for Galaxy

- No compilation at install time - binaries with their dependencies, libraries…
- Support for all operating systems Galaxy targets
- Easy to manage multiple versions of the same recipe
- HPC-ready: no root privileges needed
- Easy-to-write YAML recipes
- Community - not restricted to Galaxy
Conda recipes build packages that are published to channels.
- Recipes: independent of the progamming language in which software is written
- Support for multiple versions at the same time is needed for reproducibility
Conda Distributions
![]()
Why download Galaxy?
You need to download Galaxy if you plan to:
- Run a local production Galaxy because you want to
- Install and use tools unavailable on public Galaxies
- Use sensitive data (e.g. clinical)
- Process large datasets that are too big for public Galaxies
- Plug-in new datasources
- Develop Galaxy tools
- Develop Galaxy itself
Even when you plan any of the above sometimes you can leverage pre-configured Docker image or use Cloudlaunch
Cloud Launch
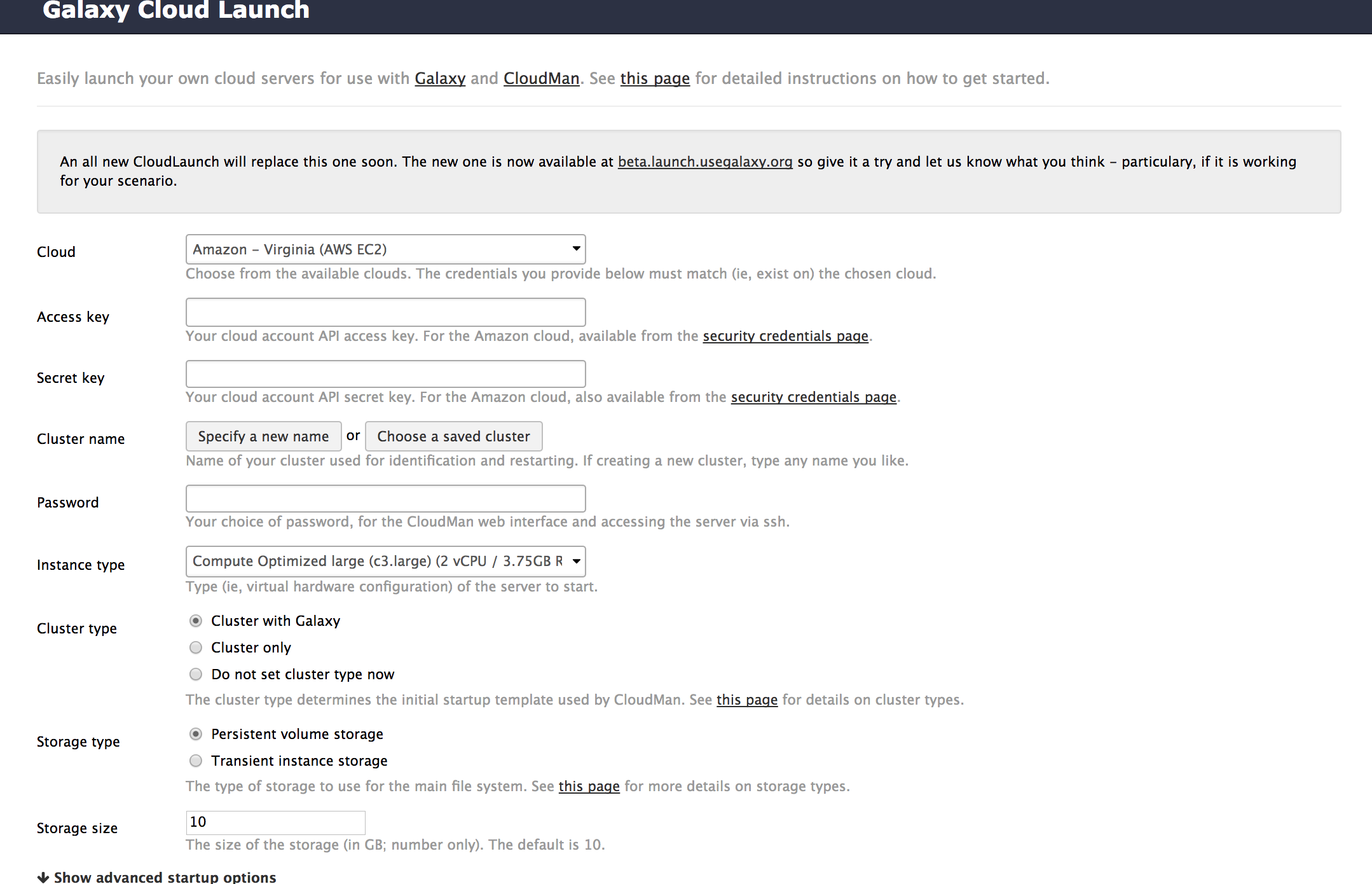
Run your own Galaxy locally
- Galaxy is open source software and can be installed on local compute infrastructure, from lab servers to institutional compute clusters
- Installing Galaxy locally is relatively easy, but
- the initial install does not include reference genomes and only has a few tools
- installing tools and genomes, setting up authentication, and connecting to institutional compute resources all takes work
- There are hundreds of local Galaxy installs around the world
- Installing tools and genomes has become much easier in recent years, and can now often be done with the Galaxy Admin GUI
- Authentication and connecting to institutional compute resources is still heavy lifting
Requirements
- Any Linux or Mac OS
- Python 2.7
Optional
- samtools (metadata etc.)
- Git code versioning system
- GNU Make + gcc to compile and install tool dependencies
- Additional requirements for shipped tools
Basic configuration
- Galaxy works out of the box with default configuration
- Most important config files are in
config/ - Galaxy often uses the files with suffix
*.sampleas declared defaults
Toolpanel management (left column)
- How the toolpanel looks like is decided in a file called
integrated_tool_panel.xml. - By default it resides in Galaxy’s root folder.
- If missing it is generated from all other tool config files during startup.
- Modify it if you want to reorder tools or move section.
The best approach for managing the new integrated_tool_panel.xml file is to allow Galaxy
to add or remove entries as manually adding or removing them will likely result in undesired behavior.
Manual changes to the file should simply be moving entries around to produce the desired arrangement of your tool panel.
Management of tool dependency installation
What resolver is going to be used for the tool dependency is determined at runtime
and prioritised in the config file dependency_resolvers_conf.xml.
<dependency_resolvers>
<tool_shed_packages />
<galaxy_packages />
<galaxy_packages versionless="true" />
<conda />
<conda versionless="true" />
<!-- other resolvers
<homebrew />
-->
</dependency_resolvers>
Galaxy Tool Shed Configuration
List of available sheds is defined in tool_sheds_conf.xml and Galaxy comes with the Main TS enabled and the Test TS disabled.
<?xml version="1.0"?>
<tool_sheds>
<tool_shed name="Galaxy Main Tool Shed" url="https://toolshed.g2.bx.psu.edu/"/>
<!-- Test Tool Shed should be used only for testing purposes.
<tool_shed name="Galaxy Test Tool Shed" url="https://testtoolshed.g2.bx.psu.edu/"/>
-->
</tool_sheds>
config/galaxy.ini
The config/galaxy.ini file contains ~300 options to be configured, grouped by sections:
- HTTP Server
- Galaxy
- Application and filtering
- Database
- Files and directories
- Tool dependencies
- Data Storage (Object Store)
- Mail and notification
- Account activation
- [Google] Analytics
- Display sites
- Next gen LIMS interface on top of existing Galaxy Sample/Request
- UI Localization
- Advanced proxy features
- Logging and Debugging
- Data Libraries
- Toolbox Search
- Users and Security: set admin user
- Beta features
- Job Execution
- ToolBox filtering
- Galaxy Application Internal Message Queue
- Galaxy External Message Queue
Files and directories
# File that can be changed by the Galaxy administrator to alter the layout of the
# tool panel. If not present, Galaxy will create it.
integrated_tool_panel_config = integrated_tool_panel.xml
Tool dependencies
# The dependency resolvers config file specifies an ordering and options for how
# Galaxy resolves tool dependencies (requirement tags in Tool XML).
# The default is
# - Tool Shed for tools installed that way
# - local Galaxy packages
# - then use Conda if available.
dependency_resolvers_config_file = config/dependency_resolvers_conf.xml
Mail and notification
# SMTP server configuration
smtp_server = None
smtp_username = None
smtp_password = None
# Datasets in an error state include a link to report the error.
# Those reports will be sent to this address.
error_email_to = None
Account activation
Require verification that a user’s email is real. You must enable SMTP first.
In galaxy.ini:
user_activation_onrequire users to click link in email before running jobs.activation_grace_periodtime (hours) that a user can ‘explore’ Galaxy before activation lockout.inactivity_box_contentmessage provided to non-activated users.- Disposable domain blacklist
blacklist_filedefines domains in XXX.YYY format that will be rejected as user emails.
UI Localization
- Show a message box under the masthead
- Append custom text to the Galaxy text in the masthead
- Custom URLs for:
- welcome_url
- logo_url
- wiki_url
- support_url
- citation_url
- Terms and conditions url
Data Libraries
In galaxy.ini:
user_library_import_dir- Directory must contain sub-directories named the same as user’s email.
- Allows users to browse and import from the given folder.
- Works well in combination with
ftp_upload_dir.
allow_library_path_paste- Admin-only, allows importing from any path that the Galaxy’s user has access to.
Users and Security
In galaxy.ini:
require_logincan be enabled to prevent anonymous access.show_welcome_with_loginshow welcome page next to login pageallow_user_creation. When False, admins must create users; often coupled withrequire_login.allow_user_dataset_purgeusers can purge (permanently delete) their datasets.api_allow_run_aslist of email addresses of API users who can make calls on behalf of other users.expose_dataset_pathusers to see the full path of datasets via the “View Details” option in the history.
Users and security - Admin
In galaxy.ini:
admin_userscomma-separated list of admin users’ emailsallow_user_deletionadmins can delete usersallow_user_impersonationadmins can become other users. Great for debugging / user assistance.master_api_keyadmin super-key allows many API admin actions without having a real admin user.
Admin Panel
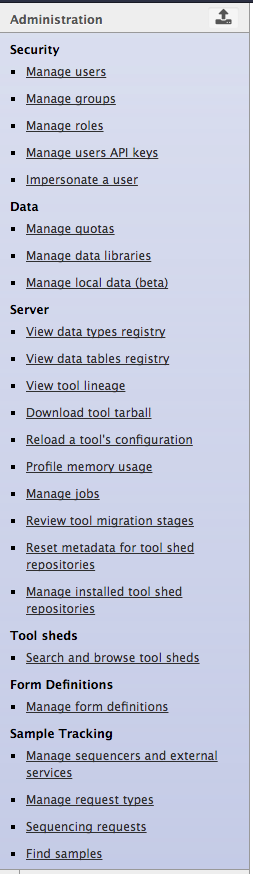
Role Based Access Control
Admin can:
- create roles (each user automatically has their own ‘private’ role)
- create groups
- assign roles to groups
- asign users to groups
- assign groups to roles
- assign users to roles
- assign permission sets to roles
- assign permission sets to groups
- Setting permissions on Libraries and Datasets
- Setting quotas