Everything in a box. Docker and Galaxy
Docker is an open platform for developing, shipping, and running applications. Galaxy is available as Docker Image, an easy distributable full-fledged Galaxy installation. Finally, Galaxy supports running tools within Docker containers.
Introduction
Linux Containers (LXC) is an operating-system-level virtualization method for running multiple isolated Linux systems (named containers) on a single control host (LXC host). It does not provide a virtual machine, but rather provides a virtual environment that has its own CPU, memory, block I/O, network, etc. space and the resource control mechanism. This is provided by namespaces and cgroups features in Linux kernel on LXC host.
Docker is an open-source engine to easily create lightweight, portable, self-sufficient containers from any application. The same container that a developer builds and test on a laptop can run at scale, in production, on VMs, OpenStack cluster, public clouds and more.
Docker can run in a VM (or not).
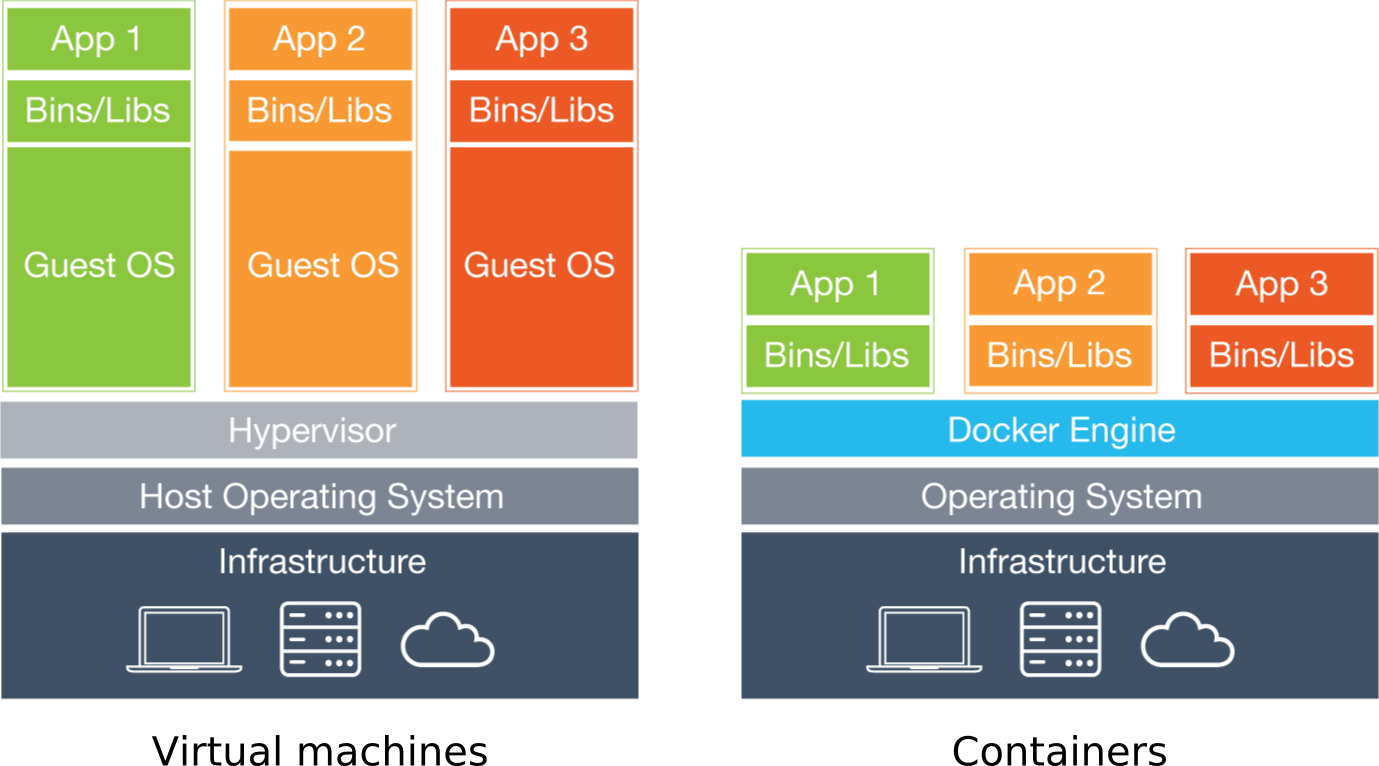
Containers and VMs are similar in their goals: to isolate an application and its dependencies into a self-contained unit that can run anywhere. The main difference between containers and VMs is in their architectural approach:
Docker features:
- Less overheads : isolation vs virtualization
- Versioning (git-like) + common base images
- Component re-use
- Nice command line based UX
- Simple reproducible builds and deployment
- Sharing (public repository)
Images and Layers:
ubuntu : 200 Mb
ubuntu + R : 250 Mb
ubuntu + matlab : 250 Mb
All three: 300 Mb
Docker: getting started
Installation
Docker is already installed on your VMs. To install it on your systems:
Search, pull, run and more
Examples:
$ docker search debian
ubuntu@workspace-new:~$ docker search debian
NAME DESCRIPTION STARS OFFICIAL AUTOMATED
debian Debian is a Linux distribution that's comp... 2129 [OK]
$ docker pull debian:jessie
jessie: Pulling from library/debian
9f0706ba7422: Pull complete
Digest: sha256:4bc62f74d246e8428be8dd3833461ba2cfd135064aed4001f3c12b87a011e30c
Status: Downloaded newer image for debian:jessie
$ docker images
REPOSITORY TAG IMAGE ID CREATED SIZE
debian jessie 62a932a5c143 4 days ago 123 MB
Running containers
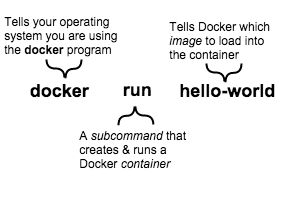
Docker runs processes in isolated containers. A container is a process which runs on a host. The host may be local or remote. When an operator executes docker run, the container process that runs is isolated in that it has its own file system, its own networking, and its own isolated process tree separate from the host. The basic docker run command:
$ docker run [OPTIONS] IMAGE[:TAG|@DIGEST] [COMMAND] [ARG...]
Example:
$ docker run --rm python:3.5 python -c "print(40 + 2)"
Unable to find image 'python:3.5' locally
latest: Pulling from library/python
357ea8c3d80b: Already exists
52befadefd24: Pull complete
3c0732d5313c: Pull complete
ceb711c7e301: Pull complete
4211bb537697: Pull complete
71f9074c0739: Pull complete
3e5349707036: Pull complete
Digest: sha256:a755ad5a30b2[...]
Status: Downloaded newer image for python:3.5
42
$ docker run --rm python:3.5 python -c "print(40 + 3)"
43
Run an interactive container
In foreground mode (the default when -d is not specified), docker run can start the process in the container and attach the console to the process’s standard input, output, and standard error. It can even pretend to be a TTY, this is what most command line executables expect
$ docker run --rm -ti python:3.5 bash
root@10d2dfedb935:/# ps
PID TTY TIME CMD
1 ? 00:00:00 bash
8 ? 00:00:00 ps
root@10d2dfedb935:/# python
Python 3.5.2 (default, Aug 9 2016, 20:58:38)
[GCC 4.9.2] on linux
>>> 40 + 2
42
>>>
root@10d2dfedb935:/# exit
Run in detached mode
To start a container in detached mode (background), use -d option. By design, containers started in detached mode exit when the root process used to run the container exits.
Example: publish nginx container port 80 on 8080 port
$ docker run -d -p 8080:80 nginx
Unable to find image 'nginx:latest' locally
latest: Pulling from library/nginx
e6e142a99202: Pull complete
8c317a037432: Pull complete
af2ddac66ed0: Pull complete
Digest: sha256:72c7191585e9b79cde433c89955547685db00f3a8595a750339549f6acef7702
Status: Downloaded newer image for nginx:latest
cc1412bff7ebf42f4173a81ee744c567b24079708ce701494faeabd645866a45
$ docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
cc1412bff7eb nginx "nginx -g 'daemon ..." 3 minutes ago Up 3 minutes 0.0.0.0:8080->80/tcp ecstatic_hypatia
$ docker exec -it ecstatic_hypatia /bin/bash
root@cc1412bff7eb:/# nginx -t
nginx: the configuration file /etc/nginx/nginx.conf syntax is ok
nginx: configuration file /etc/nginx/nginx.conf test is successful
root@cc1412bff7eb:/#
Inspect containers
$ docker ps #shows running containers
$ docker inspect #info on a container (incl. IP address)
$ docker logs #gets logs from container
$ docker events #gets events from container
$ docker port #shows public facing port of container
$ docker top #shows running processes in container
$ docker diff #shows changed files in container's FS
$ docker stats #shows metrics, memory, cpu, filsystem
Hands On! (1)
Play with biocontainers/samtools:1.3.1:
- Get the image
- Launch samtools container interactively
- Print the help page for samtools
$ docker run -t -i biocontainers/samtools:1.3.1 /bin/bash $ biodocker@4a70f09adce2:/data$ samtools --help - Launch a samtools container in detach mode
- Check if it exists and find its name
- Stop it and restart it
- Print the help page using this container
$ docker run biocontainers/samtools:1.3.1 samtools --help
Data management
Hands On! (2)
Play with biocontainers/samtools:1.3.1:
- Create locally a samtool_dir directory
- Launch an interactive container with a volume pointing at samtool_dir directory
- Add a toy sam file into the local samtool_dir directory
- Check if the existence of the file from inside the container
- Visualize the content of the toy file with samtools view commands
- Generate stats of the toy file into a toy_stat file into the local samtool_dir directory
$ mkdir -p samtools_dir
$ cd samtools_dir
$ wget https://raw.githubusercontent.com/samtools/samtools/develop/examples/toy.sam
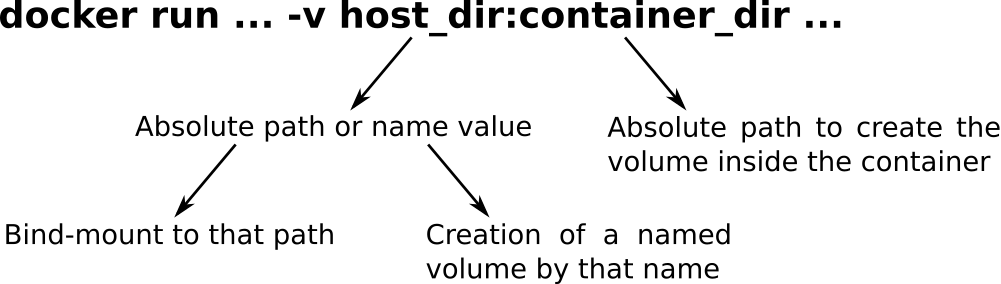
$ docker run -it -v $HOME/samtools_dir:/data biocontainers/samtools:1.3.1 /bin/bash
biodocker@cb5912bcd1be:/data$ samtools view toy.sam
r001 163 ref 7 30 8M4I4M1D3M = 37 39 TTAGATAAAGAGGATACTG * XX:B:S,12561,2,20,112
r002 0 ref 9 30 1S2I6M1P1I1P1I4M2I * 0 0 AAAAGATAAGGGATAAA *
r003 0 ref 9 30 5H6M * 0 0 AGCTAA *
r004 0 ref 16 30 6M14N1I5M * 0 0 ATAGCTCTCAGC *
r003 16 ref 29 30 6H5M * 0 0 TAGGC *
r001 83 ref 37 30 9M = 7 -39 CAGCGCCAT *
x1 0 ref2 1 30 20M * 0 0 AGGTTTTATAAAACAAATAA ????????????????????
x2 0 ref2 2 30 21M * 0 0 GGTTTTATAAAACAAATAATT ?????????????????????
x3 0 ref2 6 30 9M4I13M * 0 0 TTATAAAACAAATAATTAAGTCTACA ??????????????????????????
x4 0 ref2 10 30 25M * 0 0 CAAATAATTAAGTCTACAGAGCAAC ?????????????????????????
x5 0 ref2 12 30 24M * 0 0 AATAATTAAGTCTACAGAGCAACT ????????????????????????
x6 0 ref2 14 30 23M * 0 0 TAATTAAGTCTACAGAGCAACTA ???????????????????????
biodocker@cb5912bcd1be:/data$ samtools flagstat toy.sam
12 + 0 in total (QC-passed reads + QC-failed reads)
0 + 0 secondary
0 + 0 supplementary
0 + 0 duplicates
12 + 0 mapped (100.00% : N/A)
2 + 0 paired in sequencing
1 + 0 read1
1 + 0 read2
2 + 0 properly paired (100.00% : N/A)
2 + 0 with itself and mate mapped
0 + 0 singletons (0.00% : N/A)
0 + 0 with mate mapped to a different chr
0 + 0 with mate mapped to a different chr (mapQ>=5)
biodocker@cb5912bcd1be:/data$ samtools stats toy.sam > toy_stat
biodocker@cb5912bcd1be:/data$ exit
~/samtools_dir$ ls
toy.sam toy_stat
Creation of a new image: Dockerfile
A Dockerfile is a script, composed of various commands (instructions) and arguments listed successively to automatically perform actions on a base image in order to create (or form) a new one.
FROM ubuntu
MAINTAINER Romin Irani (email@domain.com)
RUN apt-get update
RUN apt-get install -y nginx
ENTRYPOINT [“/usr/sbin/nginx”,”-g”,”daemon off;”] # An ENTRYPOINT allows you to configure a container that will run as an executable.
EXPOSE 80
Build docker image:
$ docker build -t my_nginx_image --no-cache .
Login on Docker Hub with your credentials:
$ docker login
Login with your Docker ID to push and pull images from Docker Hub. If you don't have a Docker ID, head over to https://hub.docker.com to create one.
Username: mtangaro
Password:
Login Succeeded
Ship a Docker Image:
docker push repository_name/my_nginx_image
Docker Hub Automated build
Prerequisites: To perform Docker Hub automatic build you need a Docker Hub and on the hosted repository provider (GitHub or Bitbucket) accounts.
-
Create new Github (or bitbucket) repository.
-
Upload the Dockerfile on GitHub/Bitbucket.
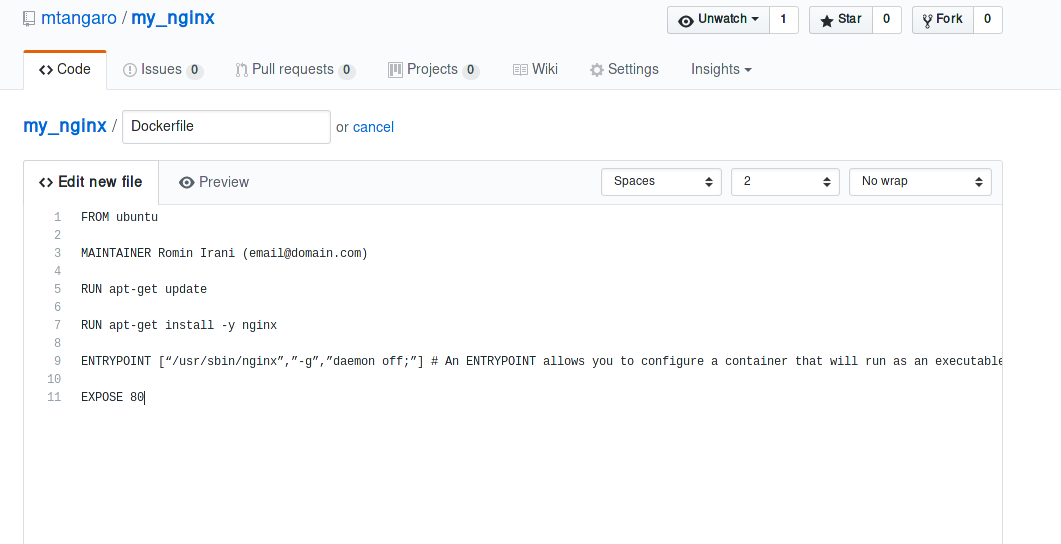
-
Login on Docker Hub and select “Create Automated Build” from Create menu.
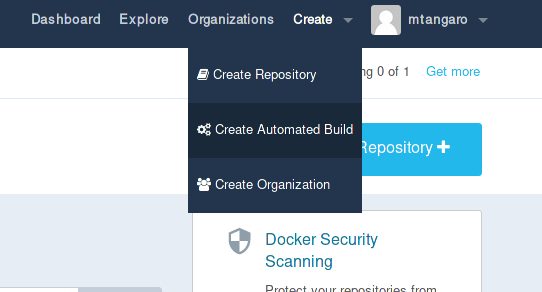
-
Link Github/Bitbucket repository to Docker Hub.
-
Select your repository with the Dockerfile.
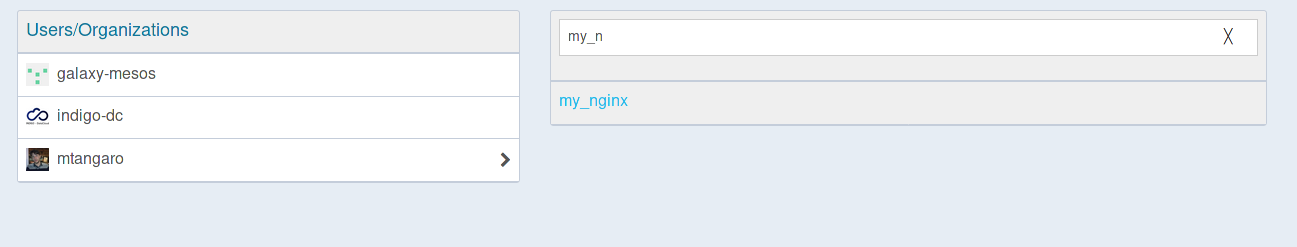
-
Trigger a build from “Build Settings” tab.
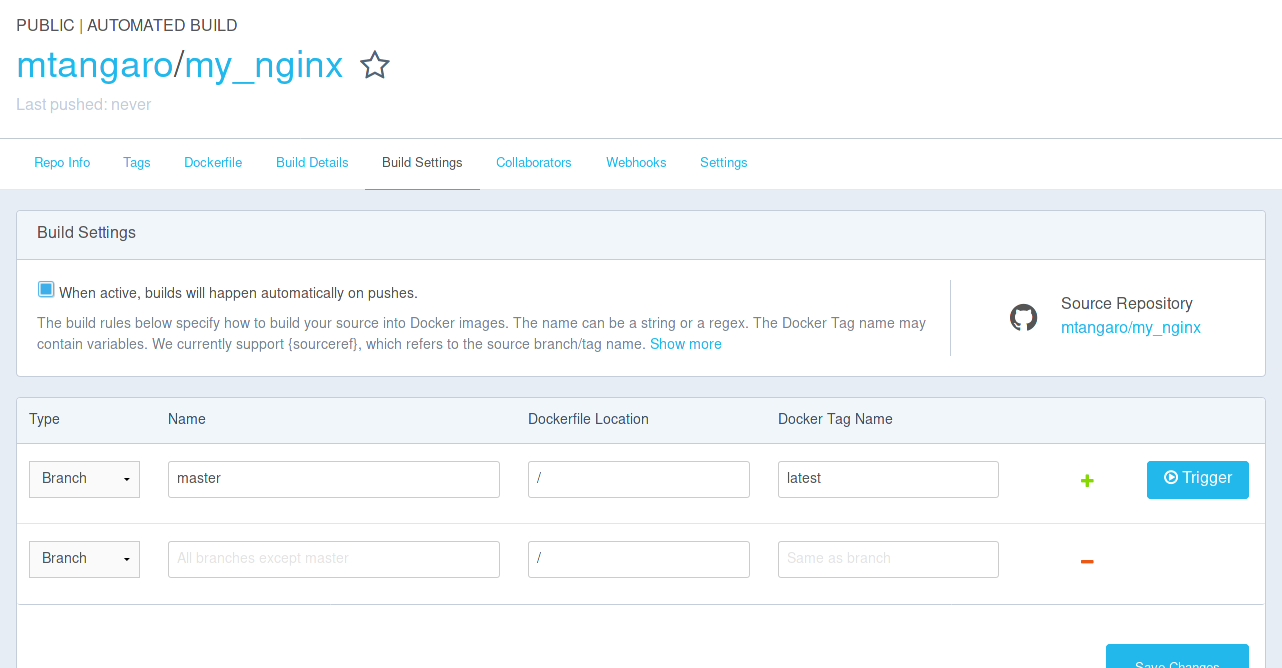
-
Pull or Run your image.
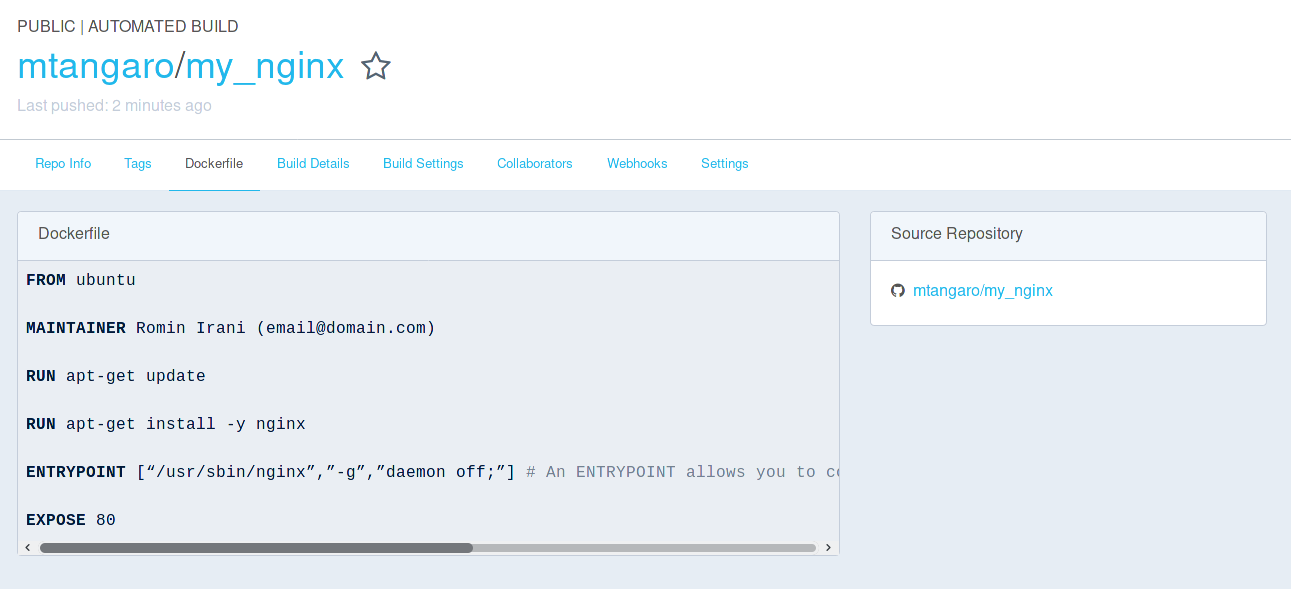
Galaxy Docker container
A Docker launching a Galaxy instance and
- FTP-Server
- Webserver
- Scheduler
- Process control UI
- ToolShed ready
- Interactive Environment ready
Link to galaxy docker stabke usage
Run Galaxy Docker:
$ docker run -d -p 9080:80 -p 9021:21 --name galaxy-stable bgruening/galaxy-stable
$ docker ps
CONTAINER ID IMAGE COMMAND CREATED STATUS PORTS NAMES
0e066ba9b720 bgruening/galaxy-stable "/usr/bin/startup" 12 seconds ago Up 11 seconds 443/tcp, 8800/tcp, 9002/tcp, 0.0.0.0:9021->21/tcp, 0.0.0.0:9080->80/tcp galaxy-stable
$ docker exec -it galaxy-stable /bin/bash
root@0e066ba9b720:/galaxy-central#
Hands On! (3)
Launch a Galaxy Docker Container and try to:
$ docker run -d -p 9080:80 -p 9021:21 --name galaxy-stable bgruening/galaxy-stable
-
Add Data
-
Register and Become an Admin
- Restart Galaxy
- Hint: edit galaxy.ini file then restart docker with
docker restart [OPTIONS] CONTAINER [CONTAINER...]
- Hint: edit galaxy.ini file then restart docker with
- Use export mounts (persistent data)
docker run -d -p 9080:80 -p 9021:21 -v $HOME/galaxy_storage/:/export/ bgruening/galaxy-stable - Trouble shooting (logging)
docker run -d -p 9080:80 -p 9021:21 -e "GALAXY_LOGGING=full" bgruening/galaxy-stable docker exec -it <container name> bashOnce connected to the container, log files are available in /home/galaxy/logs.
- Install tools …
Exteding the Docker image: Galaxy flavors
Tools that are already included in the Tool Shed, can be installed to customize Galaxy docker image (Galaxy flavor) with the following steps.
Create flavors
-
Create Dockerfile
-
Use the Galaxy Docker Image as base image and build your own extensions on top of it.
FROM bgruening/galaxy-stable MAINTAINER Björn A. Grüning, bjoern.gruening@gmail.com ENV GALAXY_CONFIG_BRAND test_flavor WORKDIR /galaxy-central RUN add-tool-shed --url 'http://testtoolshed.g2.bx.psu.edu/' --name 'Test Tool Shed' # Install Visualisation RUN install-biojs msa # Adding the tool definitions to the container ADD my_test_list.yml $GALAXY_ROOT/my_test_list.yml # Install my_tools_list RUN install-tools $GALAXY_ROOT/my_test_list.yml # Mark folders as imported from the host. VOLUME ["/export/", "/data/", "/var/lib/docker"] # Expose port 80 (webserver), 21 (FTP server), 8800 (Proxy) EXPOSE :80 EXPOSE :21 EXPOSE :8800 # Autostart script that is invoked during container start CMD ["/usr/bin/startup"] -
Supply the list of desired tools in a file (my_test_list.yml below). See this page for the file format requirements.
--- api_key: <Admin user API key from galaxy_instance> galaxy_instance: <Galaxy instance IP> tools: - name: fastqc owner: devteam tool_panel_section_label: 'Tools' install_resolver_dependencies: True - name: 'bowtie_wrappers' owner: 'devteam' tool_panel_section_label: 'Tools' install_resolver_dependencies: 'True' - Execute
docker build -t my-docker-test --no-cache . - Run your container with:
docker run -p 9080:80 my-docker-test - Open your web browser on
http://localhost:9080
List of Galaxy flavors
NCBI-Blast
ChemicalToolBox
ballaxy
NGS-deepTools
Galaxy ChIP-exo
Galaxy Proteomics
Imaging
Constructive Solid Geometry
Galaxy for metagenomics
Galaxy with the Language Application Grid tools
RNAcommender
OpenMoleculeGenerator
Workflow4Metabolomics
HiC-Explorer
SNVPhyl
GraphClust
RNA workbench
Cancer Genomics Toolkit
Integrating Docker-based tools within Galaxy
Docker is a method for wrapping up a tool along with all of it’s dependencies into a single container which can be distributed with the help of Docker-Hub. A method to run docker based tools was added to Galaxy with this pull request.
Prerequisites: enable docker to run using sudo without a password
Add docker runner to sudoers file (replace galaxy with the username you are running galaxy under):
galaxy ALL = (root) NOPASSWD: SETENV: /usr/bin/docker
Run Galaxy
-
Download Galaxy
-
Edit config/job_conf.xml adding docker runner destination, instructing Galaxy to run dockerized tools.
Construct a basic job_conf.xml with the following command.
cp job_conf.xml.sample_basic job_conf.xmlAdd a docker destination in job_conf.xml to enable running through docker:
<destinations default="docker_local"> <destination id="local" runner="local"/> <destination id="docker_local" runner="local"> <param id="docker_enabled">true</param> </destination> </destinations>More information can be found in the job_conf.xml.sample_advanced file that comes with Galaxy.
-
BWA is available via ToolShed:
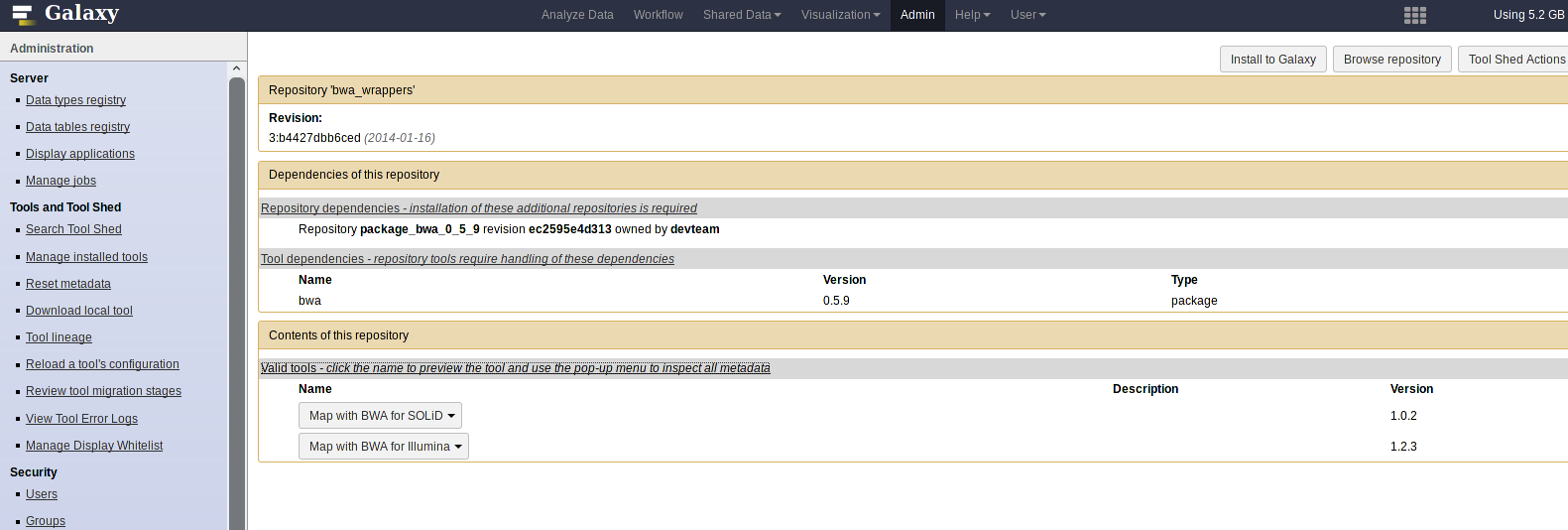
Setup bwa_wrapper.xml tool. It is located here
mkdir tools/BWA wget https://raw.githubusercontent.com/galaxyproject/tools-devteam/master/legacy/bwa_wrappers/bwa_wrapper.xml tools/BWANow, in the tool_conf.xml file please add a new section for this tool:
<section> <tool file="BWA/bwa_wrapper.xml"/> </section>Edit the xml file, changing
<requirements> <requirement type="package" version="0.5.9">bwa</requirement> </requirements>to
<requirements> <container type="docker">mtangaro/galaxy-bwa-0.5.9</container> </requirements>Remove interpreter from the command attribute by changing
<command interpreter="python"> bwa_wrapper.pyto
<command> bwa_wrapper.pyThe resulting xml wrapper is available here
Galaxy is ready!
Bwa Dockerfile
Bwa-galaxy Dockerfile is available here Docker Hub image here
sudo docker pull mtangaro/galaxy-bwa-0.5.9
bwa_wrapper.py in the docker container.
Dockerfile:
FROM ubuntu:14.04
RUN apt-get -y update
RUN apt-get install -y make build-essential zlib1g-dev python git
### install bwa
ADD https://github.com/lh3/bwa/archive/0.5.9.tar.gz /tmp/bwa.tar.gz
WORKDIR /tmp
RUN tar xvzf /tmp/bwa.tar.gz \
&& cd /tmp/bwa-0.5.9 \
&& make \
&& ln -s /tmp/bwa-0.5.9/bwa /usr/bin/
### get bwa wrapper
RUN mkdir /tmp/bwa
WORKDIR /tmp/bwa
RUN git clone https://github.com/galaxyproject/tools-devteam.git bwa_deps
RUN cp bwa_deps/legacy/bwa_wrappers/bwa_wrapper.py /usr/bin/bwa_wrapper.py
RUN chmod a+x /usr/bin/bwa_wrapper.py
Hands On! (4)
Remember: Galaxy run docker using sudo. You have to add docker runner to sudoers file.
- Everything is already available here.
Clone Galaxy repository fork, which includes all modified files:git clone https://github.com/mtangaro/galaxy.git cd galaxy git checkout Galaxy4Developers ./run.sh -
Load Input files from here: link
Reference file - fasta datatype
Forward - fastqsanger datatype
Reverse - fastqsanger datatype -
Check BWA docker on your local machine: no BWA dependencies on your machine
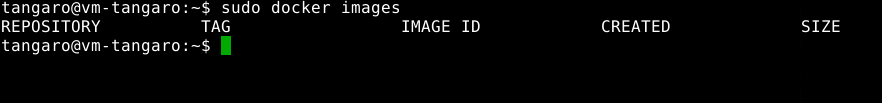
-
Configure and Run BWA job:
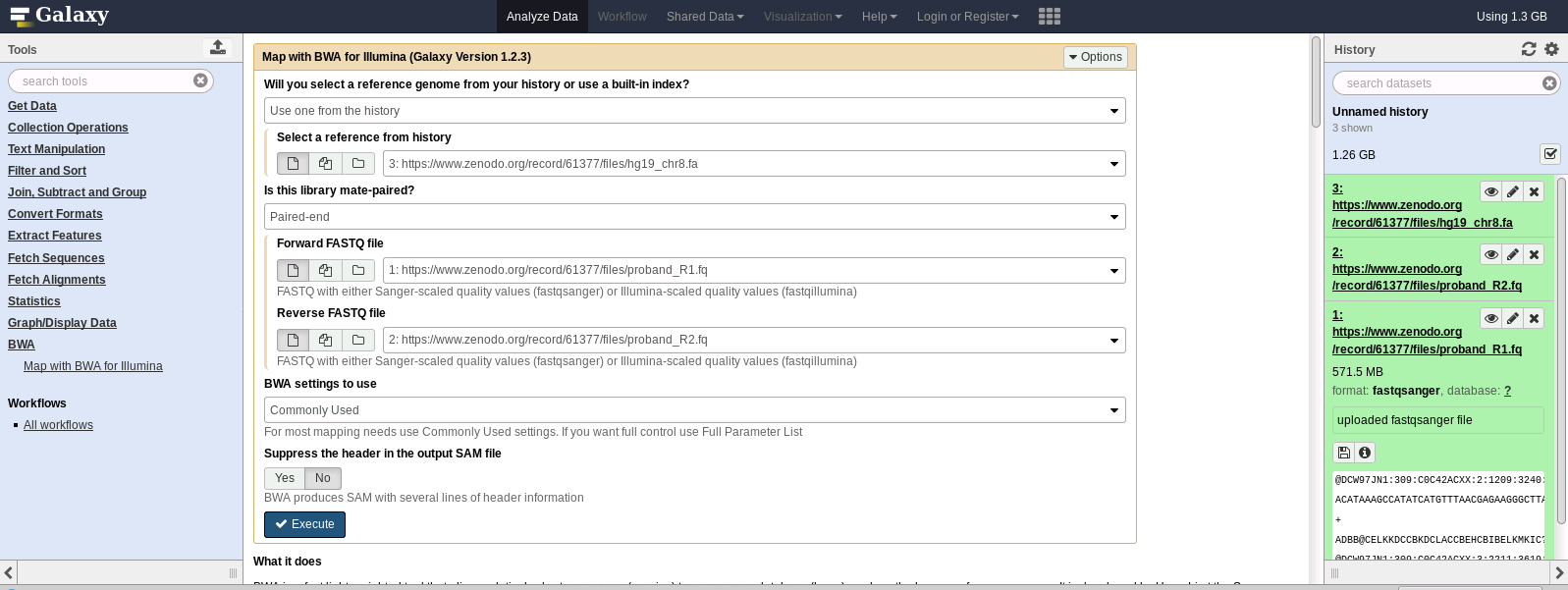
-
Run BWA. Galaxy will automatically download bwa docker image:
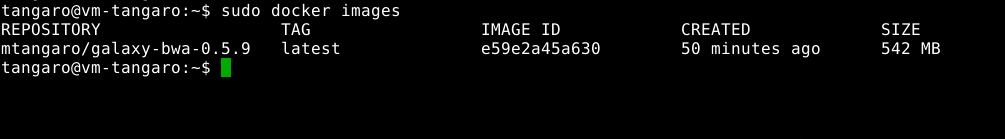
- Check results:
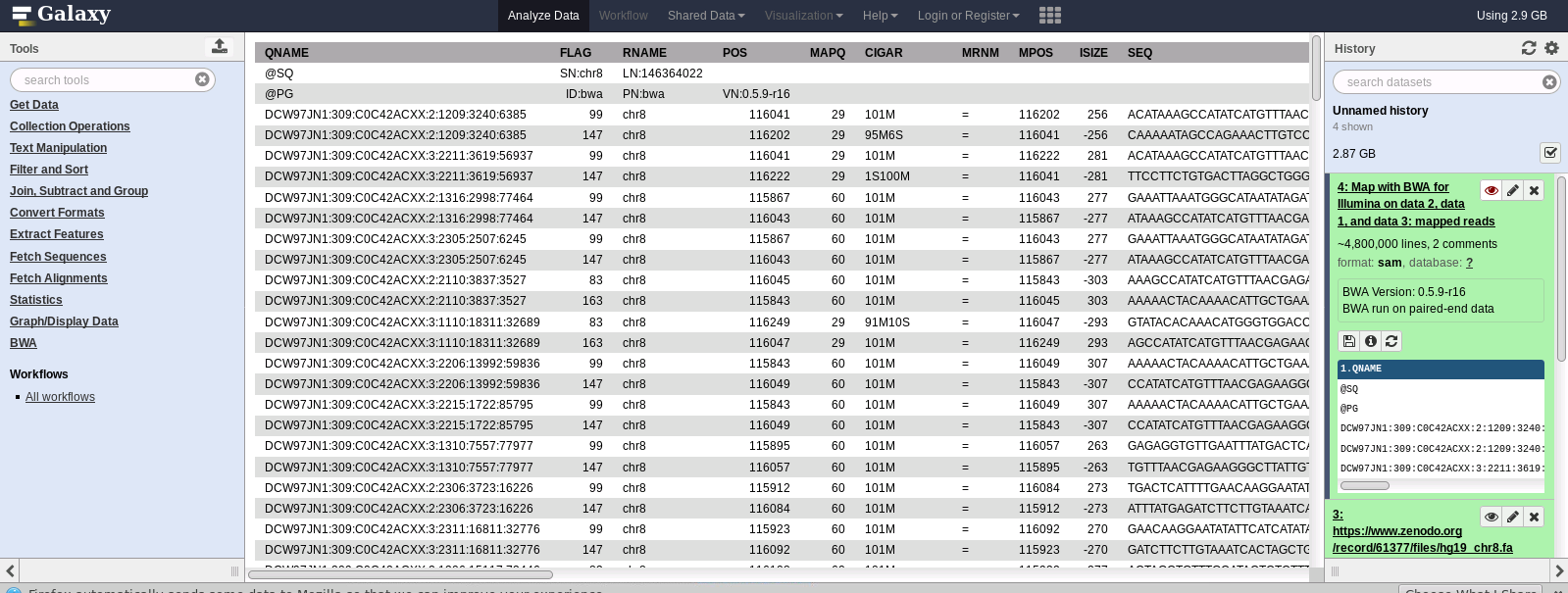
Clean your environment
Stop and remove all your containers
$ docker rm $(docker ps -a -q) -f
Remove all your images
$ docker rmi $(docker images -q)